Introduction
The prevalence and extent of antimicrobial resistance (AMR) in an animal herd is directly correlated to the total volume of antimicrobial usage (Angulo et al., 2004). In swine, where there is heavy use of antibiotics in intensive farming systems, this results in a large reservoir of antimicrobial-resistant bacteria. This is particularly so in countries like Colombia, where antimicrobials continue to be used as growth promoters in swine feed.
The prevalence and degree of AMR found in indicator bacteria in the fecal microflora of swine is a good indicator of the selection pressure of antimicrobial usage. Commensal microflora, like generic E. coli and Enterococcus spp. are typically chosen as representative indicators of AMR as they are part of the normal microflora and can acquire and disseminate resistance to pathogenic or zoonotic bacteria (Van den Bogaard et al., 2000). Thus, they can be used to compare levels of resistance between pig populations, with no resistance being the ideal goal. Medical doctors have used the analogy of not detecting resistance in indicator E. coli in healthy children to having “normal blood pressure” and “low cholesterol levels” (Lester et al., 1990). Since 2014, monitoring of AMR in indicator E. coli from food-producing animals and their food products has been mandatory under the European legislation (EFSA, 2019). There is concern that resistant bacteria may be selected in the intestinal microflora of swine contaminating food in the slaughtering process and then transferring its resistance genes to other bacteria in the human gut. In addition, the presence of E. coli on pig carcasses during slaughter is considered a reliable indicator of how good are the hygiene practices in the slaughter line (Belluco et al., 2005).
With regards to the genus Salmonella, pork has traditionally been blamed with food-borne illnesses caused by Salmonella spp. so numerous countries have established monitoring systems to report prevalence of Salmonella spp. and its antimicrobial susceptibility (Haley et al., 2012; Kadykalo et al., 2018; Kidsley et al., 2018). In recent decades, with the virtual elimination of Cysticercus cellullosae and Trichinella from pork in most developed countries, the highest concern for the safety of pork has turned to bacterial contamination, particularly with Salmonella species. Several Salmonella serovars isolated from pigs are considered important for public health, including Cholera suis, enteritidis, and typhimurium (EFSA, 2019). In addition, the genus Salmonella is also known for its potential to develop multi-drug resistance. For example, in a survey analyzing 7,788 fecal samples with an overall prevalence of 7.2% in the USA, the proportion of MDR isolates was 57.7% (Haley et al., 2012). A recent study in Colombia evaluated the overall prevalence in 21 farms across the country showing 7.6% (feces), 8.7% (rectal swabs), and a 38.1% seroprevalence (Giraldo-Cardona et al., 2019). According to another research conducted in 31 Colombian slaughterhouses, prevalence of slaughtered pigs infected with species of Salmonella spp. in lymph nodes ranged between 4.2 and 60% (Ayala-Romero et al., 2018). Because this test is a sensitive test at the individual animal level, it suggests that widespread infection is already present at the primary production site. It is necessary to conduct further studies at the moment of slaughter and in the slaughterhouse environment, as these represent a greater risk to public health.
Countries such as Canada and the United States have national surveillance programs to monitor resistance trends among specific pathogen indicator species through both active and passive means (Haley et al., 2012; Karp et al., 2017; Kadykalo et al., 2018). Passive surveillance using data from veterinary diagnostic laboratories have limitations as they may not be representative of the general bacterial populations. On the other hand, they may be important because clinically ill animals are direct targets of antimicrobial treatments and so have the greatest selective pressure.
In Colombia, there is a lack of information on the occurrence of antimicrobial resistance and, given the extended use of antimicrobials as growth promoters, studies are urgently needed to evaluate the current situation. Therefore, the aim of this study was to investigate the state of AMR in generic E. coli and Salmonella spp. isolated from cecal contents of pigs submitted to a veterinary diagnostic laboratory in Colombia from 2016 to 2019.
Materials and Methods
Results of antimicrobial susceptibility test were obtained at the Laboratorio de Diagnóstico Veterinario of the Universidad de Antioquia from fecal samples of swine cases submitted between 2016 and 2019. Susceptibility testing was conducted using the Kirby-Bauer disk diffusion method according to the Clinical and Laboratory Standards Institute guidelines for antimicrobial zone diameter breakpoints (CLSI, 2018). The antimicrobials (Oxoid, Hampshire, United Kingdom) tested were: amoxicillin (10 ug), ceftiofur (30 ug), ciprofloxacin (5 ug), florfenicol (30 ug), gentamicin (10 ug), sulfamethoxazol/trimethoprim (25 ug), tetracycline (30 ug), enrofloxacin (5 ug), doxycyclin (30 ug), neomycin (30 ug), and tylosin (30 ug). E. coli strain ATCC 25922 was used as the CLSI quality control strain for antimicrobial susceptibility testing. Isolates showing resistance to three or more antimicrobial classes were classified as multidrug-resistant (MDR) as defined by a joint group of the European Centre for Disease prevention and Control and the Center for Disease Control and Prevention of the USA (Magiorakos et al., 2012).
On the day of arrival, 10 g of fecal material was suspended in 7 mL of 0.1% sterile peptone water and mixed before 1 mL of the mixture was extracted and centrifuged. The homogenate was plated onto MacConkey agar (Oxoid, Thermofisher Scientific, Hampshire, United Kingdom) and incubated at 37 °C for 24 h.
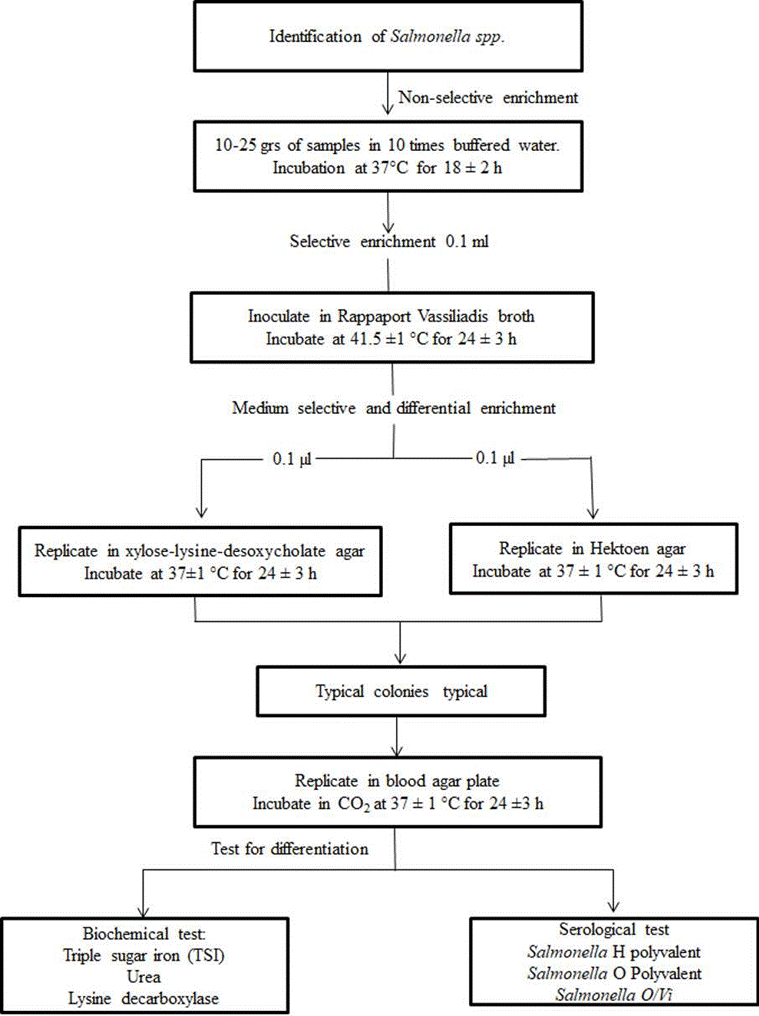
Lactose positive presumptive E. coli colonies were subcultured onto sheep blood agar (Oxoid, Themofisher Scientific, Hampshire, United Kingdom) and incubated at 37 °C for 24 h. The isolates on nutrient agar were subjected to an indole test for E. coli identification. For Salmonella spp. isolation and colony identification, the steps followed the sequence shown in Figure 1.
Statistical analysis
The data were entered into Excel 2010 and descriptive statistics used to assess frequency of resistance. Since few results were classified as intermediate (neither susceptible nor resistant to the antimicrobial), for discussion purposes they were reclassified in the resistant category.
Results
A total of 557 fecal samples were submitted be- tween 2016 and 2019 from which 112 and 192 E. Coli and Salmonella spp. colonies were isolated, respectively. Results of antimicrobial resistance and susceptibility patterns of the isolated E. coli and Salmonella spp. colonies are presented in Tables 1 and 2. Not all antibiotics were tested on all strains. For E. coli, the percentage resistance, in order of decreasing prevalence was: tetracycline (100%), sulfamethoxazol-trimethoprim (97.5%), ampicillin (95.4%), amoxycillin (86.4%), enrofloxacin (82.6%), tylosin (82.1%), doxycycline (59%), neomycin (50%), ciprofloxacin (45.5%), ceftiofur (35%), gentamicin (30%), tilmicosin (29%), fosfomycin (12.5%). With regards to Salmonella spp., the decreasing order of resistance prevalence was: tetracycline (100%), tylosin (89%), florfenicol (84.4%), tilmicosin (80%), doxycyclin (76.5%), enrofloxacin (72%), sulfamethoxazol-trime-thoprim (68.5%), ampicillin (66.7%), amoxicillin (63.6%), ciprofloxacin (29.5%), ceftiofur (27.4%), neomycin (17%), gentamicin (16%), fosfomy- cin (14%). Multi-drug resistance to three or more classes of antimicrobials was observed in 68.7% (77/112) of the E. coli isolates, and it was 70.3% (135/192) for the Salmonella spp. isolates.
Table 1 Susceptibility of generic E. coli isolated from 112 swine fecal samples to different antimicrobial agents.
| Resistant | Intermediate | Susceptible | |||||
|---|---|---|---|---|---|---|---|
| Antimicrobial agent | N | n | % | n | % | n | % |
| Enrofloxacin | 86 | 51.0 | 59.3 | 20.0 | 23.3 | 15.0 | 17.4 |
| Sulfamethoxazol- Trimethoprim | 79 | 77.0 | 97.5 | 0.0 | 0.0 | 2.0 | 2.5 |
| Ciprofloxacin | 68 | 31.0 | 45.6 | 0.0 | 0.0 | 37.0 | 54.4 |
| Gentamicin | 67 | 19.0 | 28.4 | 1.0 | 1.5 | 47.0 | 70.1 |
| Florfenicol | 63 | 47.0 | 74.6 | 4.0 | 6.3 | 12.0 | 19.0 |
| Amoxycillin | 59 | 49.0 | 83.1 | 2.0 | 3.4 | 8.0 | 13.6 |
| Ceftiofur | 57 | 15.0 | 26.3 | 5.0 | 8.8 | 37.0 | 64.9 |
| Fosfomycin | 48 | 6.0 | 12.5 | 0.0 | 0.0 | 42.0 | 87.5 |
| Tetracycline | 46 | 46.0 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Tylosin | 28 | 22.0 | 82.1 | 0.0 | 0.0 | 6.0 | 17.9 |
| Tilmicosin | 24 | 7.0 | 29.2 | 0.0 | 0.0 | 17.0 | 70.8 |
| Ampicillin | 21 | 20.0 | 95.2 | 0.0 | 0.0 | 1.0 | 4.8 |
| Neomycin | 20 | 9.0 | 45.0 | 1.0 | 5.0 | 10.0 | 50.0 |
| Doxycycline | 17 | 7.0 | 41.2 | 3.0 | 17.6 | 7.0 | 41.2 |
Table 2 Susceptibility of Salmonella spp. isolated from 192 swine fecal samples to different antimicrobial agents.
| Resistant | Intermediate | Susceptible | |||||
|---|---|---|---|---|---|---|---|
| Antimicrobial agent | N | n | % | n | % | n | % |
| Enrofloxacin | 162 | 66.0 | 40.7 | 51.0 | 31.5 | 45.0 | 27.8 |
| Sulfamethoxazol- Trimethoprim | 143 | 97.0 | 67.8 | 1.0 | 0.7 | 45.0 | 31.5 |
| Florfenicol | 141 | 117.0 | 83.0 | 2.0 | 1.4 | 22.0 | 15.6 |
| Ciprofloxacin | 132 | 22.0 | 16.7 | 17.0 | 12.9 | 93.0 | 70.5 |
| Gentamicin | 101 | 12.0 | 11.9 | 4.0 | 4.0 | 85.0 | 84.2 |
| Fosfomycin | 100 | 12.0 | 12.0 | 2.0 | 2.0 | 86.0 | 86.0 |
| Amoxicillin | 99 | 61.0 | 61.6 | 2.0 | 2.0 | 36.0 | 36.4 |
| Ceftiofur | 62 | 13.0 | 21.0 | 4.0 | 6.5 | 45.0 | 72.6 |
| Neomycin | 53 | 6.0 | 11.3 | 9.0 | 17.0 | 44.0 | 83.0 |
| Ampicillin | 39 | 26.0 | 66.7 | 0.0 | 0.0 | 13.0 | 33.3 |
| Tetracycline | 39 | 39 | 100.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Tylosin | 27 | 24.0 | 88.9 | 1.0 | 3.7 | 2.0 | 7.4 |
| Cefalothin | 23 | 7.0 | 30.4 | 2.0 | 8.7 | 14.0 | 60.9 |
| Doxycycline | 17 | 12.0 | 70.6 | 1.0 | 5.9 | 4.0 | 23.5 |
| Tilmicosin | 10 | 7.0 | 70.0 | 1.0 | 10.0 | 2.0 | 20.0 |
Discussion
The aim of this study was to investigate the frequency of AMR among E. coli and Salmonella spp. isolates from pig feces submitted to the Laboratorio de Diagnóstico Veterinario of the Universidad de Antioquia, Colombia. A major finding was the extremely high level of resistance (70-100%) in both bacteria to tetracycline, doxycycline, sulfomethoxazol-trimethoprim, florfenicol, tylosin and enrofloxacin. The overall frequency of E. coli resistance to the antimicrobials with the highest resistance levels was even higher to that reported by the southern and eastern European countries in 2017 (EFSA, 2019). For example, when comparing the results of our study to Spain, the overall resistance in decreasing order of frequency was: tetracycline (100 vs 88.8%), sulfamethoxazole (100 vs 63.6%) trimethoprim (100 vs 60.6%), ampicillin (95.4 vs 77.1%), and ciprofloxacin (45.5 vs 44.7%). In Spain, 60% of the E. coli isolates were MDR, being one of the countries in Europe with the largest number of MDR E. coli, with an overall mean of 34.9% for all the EU countries, ranging from 3.3% in Norway up to 82.5% in Cyprus. In our study, MDR for E. coli was 68.7% (higher than the 60% for Spain). When this resistance pattern is compared with the amount of antimicrobials used in pigs for the most common clinical conditions in Europe, a clear association becomes apparent. For respiratory diseases, 88% of the treatments mentioned were non-critically important antimicrobials (CIA), mostly tetracyclines (47%) and penicillins (21%) (De Briyne et al., 2014). These were followed by macrolides such as tylosin (10%), and potentiated sulphonamides (8%). For diarrhea, polymyxin (mostly colistin) was reported to be used 30% of the times, followed by macrolides (10%), potentiated sulfonamides (9%), fluoroquinolones (8%), and tiamulin (7%).
There are no similar studies in Colombia reporting the overall use of antimicrobials in pigs, but considering that resistant trends were closely similar to those in Spain, it is likely that the type of antimicrobials used for the most common clinical conditions are similar. In addition, Colombian legislation does not compel feed companies to declare in the label of their products the type of growth promoters used; thus, it is likely that antimicrobials with the highest level of resistance (i.e., tetracycline, sulfamethoxazole-trimethoprim, tylosin) are used as growth promoters. In a systematic review of the most common isolated resistant pathogens from different food animals in Colombia, E. coli and Salmonella spp. were the most common bacteria showing resistance to beta-lactams, macrolides (tylosin) and tetracycline (Arenas and Moreno-Melo, 2018). In our findings, the observed high frequency of resistance to amoxicillin (86.4% for E. coli) could be explained by high usage of beta-lactams and also from co-selection of genes encoding-lactamase production, which are often located on the same plasmid as the genes for tetracycline resistance (Nijsten et al., 1996). Before colistin was considered a CIA by WHO (WHO, 2017), the relative mention of use of CIA to non-CIA in pigs was about 20% in Europe, with macrolides and fluoroquinolones accounting for the main groups suggested to be restricted in pigs in those countries that still used them widely (De Briyne et al., 2014).
With regards to Salmonella spp., the resistance pattern observed closely mirrored that for E. coli, but it tended to have lower resistance to some antimicrobials. When comparing our results to those of Spain in 2017, the overall frequency in decreasing order of frequency was: tetracycline (100 vs 75%), sulfamethoxazole (68.5 vs 72%), ampicillin (66.7 vs 67.1%), ciprofloxacin (29.5 vs 20.7%), and gentamicin (16 vs 9.8%; ESFA, 2019). In their report, an overall 51.3% of Salmonella spp. isolates were MDR, but in our study 70.3% of the isolates were MDR, making them even more dangerous from a potential zoonotic standpoint than susceptible strains. A recent study in Colombia determined Salmonella spp. prevalence and antimicrobial resistance pattern from healthy pig fecal and swab samples, finding not so high resistance against some of the antimicrobials that we tested, such as ampicillin (33 vs 66.7%), ciprofloxacin (11.1 vs 29.5%), and trimethropim-sulfametoxazole (50 vs 68.5%; Giraldo-Cardona et al., 2019). The higher resistance found in our study could be explained by the fact that our samples did not represent the general swine population and most samples submitted to the diagnostic laboratory probably came from clinically ill animals exposed to antimicrobial treatments.
The emergence of antibiotic resistance in non-typhoidal Salmonella spp. associated with antimicrobial use in pig production has been well documented. Campos et al., (2019) highlighted the contribution of different drivers to the overall resistance burden. Their review found that acquisition of resistance mechanisms to antibiotics with the highest resistance in our study (amoxicillin, tetracycline, sulphonamides) was relevant for their potential role in co-selection of pig-related MDR Salmonella clones and further transmission to humans. They concluded that “the pig production setting can be a relevant reservoir of successful and worldwide emergent MDR pig- related Salmonella serotypes/clones, enriched with different features (e.g., metal/biocides tolerance genes) besides genes conferring resistance to critical antibiotics, which might spread to humans through the food chain”.
The high bacterial resistance observed in pig fecal samples calls for a more rational usage of antimicrobials and the implementation of policies to safeguard their therapeutic efficacy and minimize public health risk. A national surveillance program should also be started to monitor antimicrobial resistance as part of the FAO, WHO, and OIE global initiatives. Colombia should observe WHO recommendations and restrict the growth promotion use of medically important antimicrobials in food-producing animals (Aidara-Kane et al., 2018). Guidelines on the prudent use of antimicrobials have been implemented in various swine producing countries (Magnusson et al., 2019; Cutler et al., 2020) and it is of utmost necessity to implement them in Colombia. Those guidelines should provide recommendations on the most common antimicrobials prescribed by veterinarians, starting with ways to prevent and control the underlying causes of disease by adopting good management and husbandry practices (it has long been recognized that the best response to antimicrobials is obtained under poor hygiene conditions).
A proof that guidelines and control strategies work is that countries such as Denmark and Australia are now successfully raising pigs with little reliance on antimicrobials.
In conclusion, our results indicate there is a high level of multi-drug resistance (MDR) in Colombia. It is necessary to implement a nationwide antimicrobial resistance monitoring program in Colombia together with antimicrobial prescribing guidelines for pigs. The indiscriminate use of antimicrobials for growth promotion in the swine industry is clearly promoting widespread resistance and should be discontinued.