Introduction
Helicobacter pylori infection has importance in pathogenesis of ulceration and gastric cancer, among other extra-digestive diseases in humans (Figura et al., 2010). This bacteria is gram-negative, microaerophilic, flagellated, pleomorphic (although its usual shape is spiral; Hermanns et al., 1995), oxidase, catalase, and urease positive (Montgomery et al., 1988). Studies on Helicobacter spp. conducted in ferrets, leopards, primates, calves (Hermanns et al., 1995), sheep, pigs (Barbosa et al., 1995), cats and dogs (Jalava et al., 1998; Neiger and Simpson, 2000) showed high gastric presentation. Inoculation of H. pylori in animal models has developed the same pattern of chronic active gastritis as in humans (Lee, 1998). Cardona et al. (2009) described chronic inflammatory patterns in gastric mucosa of equines with presence of Helicobacter spp. determined by rapid urease test and histological evaluation. Although several species, such as H. felis, H. heilmannii, H. bizzozeronii, H. bitis, H. salomonis, H. rappini, have been isolated besides H. pylori in animals, there is little information on equines regarding the relationship between this infection and gastric disease. There is little information on ways of infection, transmission and/or zoonotic potential of Helicobacter spp. in equines.
H. pylori diagnosis can be performed with invasive (direct) or noninvasive (indirect) methods. Invasive methods require trans- endoscopic collection of biopsies for identification of the bacterium, such as rapid urease testing, cytology, histopathology, cultures and PCR. Non-invasive methods depart from the indirect demonstration of the presence of the bacteria, such as the labeled urea breath test and serological tests.
The presence of this bacteria in the gastric surface of equines -with other microorganisms and bacterial metabolites- has been proposed as a cause for equine gastric ulcer syndrome (EGUS). Helicobacter pylori and Helicobacter equorum have been isolated in ulcerated and healthy equines with controversial participation in EGUS physiopathology in Europe and North America, where there is high incidence and prevalence of this syndrome (Scott et al., 2001; Bezdekova and Futas, 2009; Moyaert et al., 2009).
Due to its high transmissibility, possible zoonotic potential, association between Helicobacter spp. and gastric pathologies in humans, and the presence of EGUS (specially equine gastric glandular disease-EGGD), it is relevant to understand the participation of this bacterium in gastric mucosa alterations in the Colombian Creole horse and the homology between the species infecting both humans and equines.
Therefore, the aim of this study was to assess the presence of Helicobacter spp. DNA in equine gastric mucosa and dental tartar and evaluate the relationship between presence of these bacteria and gastric lesions. We hypothesized no relationship exists between Helicobacter spp. colonization and EGGD. We also assumed that Helicobacter spp. is present in the dental tartar.
Materials and Methods
Ethical considerations
This study was approved by the Ethics Committee in Animal Experimentation of the University of Antioquia, Colombia (endorsement: June 2014).
Animals
A convenience sample from 30 equines taken at a slaughterhouse in Rionegro municipality, province of Antioquia (Colombia) was used. The facilities are located at coordinates 6°09′12″N 75°22′27″W, with 2,080 m.a.s.l. and the average temperature is 18.5 °C.
Evaluation of the gastric surface
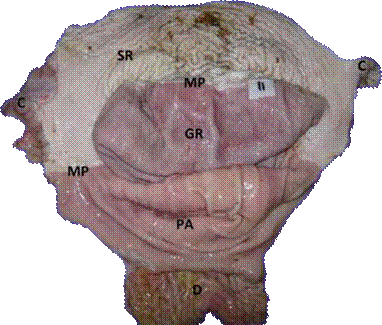
The stomachs obtained post-mortem were opened and washed with distilled water, identified numerically and later photographed. The lesions found in the glandular regions, pyloric antrum, squamous area (Margo plicatus) and cardial area were classified according to the system recommended by Sykes et al. (2015; Figure 1).
Sample collection
Stomach and head of each individual, previously prepared and numbered, were sampled. Fragments of glandular gastric mucosa (fundus) of approximately 1 cm2 were obtained using a sterile scalpel blade for each animal. Tartar samples were obtained by friction with a curette (sanitized with 2% glutaraldehyde between individuals) on the enamel surface of the upper incisors. Subsequently, both were submerged in a 2.5 ml cryovial with sterile PBS 1X for freezing (-20 ºC). These samples were directly taken at the slaughterhouse, from horses starved for at least eight hours.
Total DNA extraction
Total DNA extraction was performed with a commercial extraction kit (Wizard® Genomic DNA Purification Kit; Madison, WI, USA) added with proteinase K (60 µl per sample) for the processing of the stomach samples. Dental tartar samples were subjected to extraction with columns (Qiagen® DNeasy Blood & Tissue Kit; Hilden, Germany). Once total DNA was extracted, electrophoresis was performed by adding 1 µl of loading buffer to 1 µl of each sample on a 2% agarose gel in order to evidence the extracted product. The extracted DNA was stored in a cryovial for freezing (-20 ºC).
Amplification of the encoding gene of the Helicobacter spp. 16s rRNA region
A final-point PCR was carried out using primers for amplification of a 251 bp segment of the gene encoding the 16s rRNA, whose sequence was: HelF (forward) 5’-CGTGGAGGATGAAGGTTTTA-3’ and HelR2 (reverse) 5’-AATTCCACCTACCTCTCCC-3’ (Recordati et al., 2007).
A commercial mix was used for PCR (GoTaq Green Master Mix®; Madison, WI, USA) (10.5 µl per reaction), added with 0.5 µl of each primer (initiator and reverse), 10 µM, 4.5 µl of deionized sterile water and 2 µl of DNA (final volume per reaction: 18 µl). The PCR conditions were as follows: 5 minutes at 94 ºC, followed by 40 cycles of denaturalization at 94 ºC for 30 seconds; alignment of primers at 45 ºC for 30 seconds, and extension to 72 ºC for 30 seconds, with a final extension step at 72 ºC for 7 minutes. Helicobacter pylori DNA, donated by the Gastrohepathology laboratory of the Medicine School at Universidad de Antioquia (Figure 2), was used as positive control.
Amplification of the encoding gene of the Helicobacter pylori VacA region
Primers for the H. pylori VacA region were used since it is considered to be a species’ own virulence factor. The detection of a gene sensitive for detecting the microorganism was also considered. The PCR results that were identical to the positive control (encoding gene of the 16s rRNA region) were subjected to a new PCR to determine the presence of H. pylori.
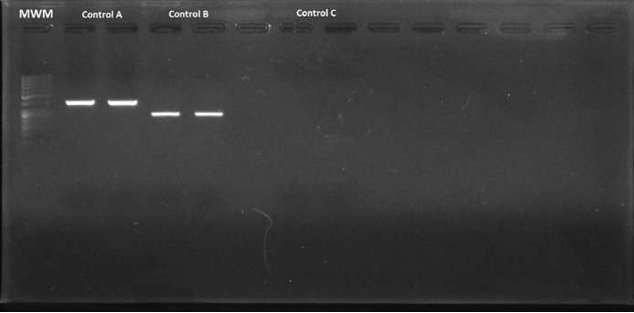
Figure 2 Electrophoresis in agarose gel at 2% of the amplified fragments of the coding gene for the Helicobacter pylori 16s rRNA region (positive control). MWM = Molecular weight marker; Control A = Amplified fragments of primers C98 and C97 (16s rRNA) of the duplicate positive control; Control B = Amplified fragments of primers HelF and HelR (16s rRNA) of the duplicate positive control; Control C = Duplicate negative control.
To this end, the VacA gene was typified in the samples by using VacA primers (s) (initiator) 5’-ATG GAA ATA CAA CAA ACA CAC 3’ (reverse) 5’ CTG CTT GAATGC GCC AAA C 3’; and VacA (m) (initiator) 5’ CAA TCT GTC CAA TCA AGC GAG 3’ (reverse) 5’ GCG TCT AAA TAA TTC CAA GG 3’. The PCR conditions for VacA “s” and “m” were as follows: 2 minutes at 92 ºC, followed by 35 cycles of denaturalization at 94 ºC for 1 minute, alignment of primers at 55 ºC for 1 minute, and extension to 72 ºC for 1 minute.
Sequencing
Sequencing was performed in Macrogen™ laboratories (Korea). Sequences were analyzed using BLAST (Basic Local Alignment Search Tool; http://blast.ncbi.nlm.nih.gov) to compare the nucleotide sequences from the PCR products of the amplified region with similar sequences reported in the GenBank.
Results
Gastric tissues and dental tartar samples required maceration prior to the extraction protocol. All the stomach samples evidenced DNA extracted by electrophoresis in 2% agarose gel. In contrast, only some of the tartar samples evidenced DNA through electrophoresis. It was possible to amplify the encoding fragment for the 16s rRNA region. However, for the VacA gene (s and m) no amplification was found, even though the positive control worked properly. The samples (stomach and tartar) from which fragments were amplified were considered Helicobacter-positive. These fragments have gel diffusion compatible with the sequence of the coding gene of the Helicobacter spp. 16s rRNA region. 23.3% (7/30) of the stomach samples were positive, while 10% (3/30) of tartar samples were positive.
The 3 or 4-degree lesions were evidenced in the Margo plicatus, or cardial region, and some of those injured stomachs were Helicobacter- positive (4/7). The remaining positive results corresponded to stomachs classified with lesions 0 - 2 (Table 1).
The sequences obtained were not clear and confirmation of Helicobacter spp. DNA was difficult in the amplified fragment. Fragments of the clearer sequences were compatible with H. heilmannii in 25 nucleotides. The positive control sequence was 100% compatible with H. pylori in both gastric mucosa and tartar.
Table 1 Classification of gastric lesions (according to Sykes et al., 2015) and result of the PCR for each region, per individual.
| Sample/ Horse | Glandular region | Pyloric antrum | Squamous region (Margo plicatus) | Cardial region | PCR (Stomach) | PCR (Tartar) |
|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 0 | 0 | + | |
| 2 | 2 | 0 | 0 | 4 | ||
| 3 | 1 | 0 | 0 | 0 | + | |
| 4 | 0 | 0 | 0 | 0 | ||
| 5 | 2 | 0 | 0 | 0 | + | |
| 6 | 1 | 0 | 0 | 0 | ||
| 7 | 1 | 0 | 4 | 0 | + | |
| 8 | 0 | 0 | 0 | 0 | ||
| 9 | 2 | 0 | 0 | 0 | ||
| 10 | 1 | 0 | 3 | 0 | + | |
| 11 | 0 | 0 | 0 | 0 | ||
| 12 | 0 | 2 | 0 | 0 | ||
| 13 | 1 | 0 | 0 | 0 | ||
| 14 | 0 | 0 | 0 | 0 | ||
| 15 | 1 | 1 | 0 | 0 | ||
| 16 | 2 | 0 | 0 | 0 | ||
| 17 | 0 | 1 | 0 | 0 | + | |
| 18 | 1 | 0 | 0 | 0 | + | |
| 19 | 1 | 0 | 0 | 4 | + | |
| 20 | 1 | 0 | 4 | 4 | ||
| 21 | 1 | 0 | 4 | 4 | ||
| 22 | 0 | 0 | 0 | 3 | + | |
| 23 | 1 | 0 | 0 | 4 | ||
| 24 | 1 | 3 | 0 | 0 | ||
| 25 | 1 | 0 | 0 | 0 | + | |
| 26 | 0 | 1 | 0 | 0 | ||
| 27 | 1 | 0 | 0 | 0 | ||
| 28 | 2 | 0 | 3 | 0 | ||
| 29 | 0 | 0 | 0 | 0 | ||
| 30 | 1 | 0 | 0 | 3 |
+ = Amplification of the encoding gene for the Helicobacter spp. 16s rRNA region.
Discussion
According to our results, it was not possible to establish a relationship between lesions in the squamous mucosa (equine squamous gastric disease-ESGD) or glandular mucosa (EGGD) and the presence of Helicobacter spp. This confirms that, as has been described in other horse breeds, infection by Helicobacter spp. and ESGD or EGGD are not related.
In addition, sequencing and verification of the species in GenBank detected a homology of 25 nucleotides as H. heilmannii. This specie has been reported as a commensal in horses (Perkins et al., 2012; Dong et al., 2016). Other researchers (Husted et al., 2010) reported that Helicobacter does not appear in the gastric microbiome of slaughtered horses; not even in lesioned gastric tissue. To the best of our knowledge, this is the first report evidencing the presence of Helicobacter spp. in dental tartar in horses. On the other hand, UREI gene of Helicobacter has been used to detect gastric infection in horses and, in contrast with our results, could found developing infections (Hepburn, 2004).
Some researchers reported sample preserva- tion in absolute ethanol to ensure integrity of the bacterium (Contreras et al., 2007). In contrast, the present study preserved the samples in ster- ile PBS 1X; this difference could have affected the quality of the extracted DNA, since loss of tissue integrity in some samples was evidenced after cryopreservation, which was reflected in DNA damage evidenced in electrophoresis by the appearance of a great number of artifacts. Because glutaraldehyde 2% was used to sanitize the curette, it might have caused the inconsistent sequencing results (Churro et al., 2015). This could imply loss of positive results.
The total DNA extraction protocol was modified by adding proteinase K to achieve complete tissue dehiscence. The PCR protocol used was taken from a study on gastric mucosa and dental tartar in dogs; however, even though the described conditions were replicated, numerous bands by sample of DNA subjected to electrophoresis were obtained, so such protocol also suffered a temperature adjustment of primer alignment.
Due to the method used to obtain stomach samples post-mortem it was possible to process the totality of them (including all stomach layers from the mucosa to the serosa), ensuring detection of the bacterium in any of the layers. This differs from studies conducted from biopsy samples by endoscopy, where the fragments obtained only sometimes include mucus and epithelial cells of the mucous layer.
As described by Contreras et al. (2007), the appearance of DNA compatible with Helicobacter spp. was more frequent in stomachs with mild or non-existing lesions, whereas the majority of those characterized as deep lesions did not correspond to positive samples for Helicobacter spp. This can happen because the loss of tissue integrity does not offer favorable conditions for the microorganism to remain viable, and therefore it is presumed that this microorganism moves to healthy tissue, facilitating its detection there. Consequently, establishing the relationship between presence of Helicobacter spp. and gastric lesions is a complex issue.
Our results contrast with those conducted in humans since they have shown 100% association in both samples, in bacterial plaque or tartar and in gastric mucosa (Scarano et al., 2005), while the present study did not find any relationship between dental tartar and gastric mucosa results in equines.
In conclusion, 23.3% of stomach and 10% of tartar samples amplified for the gene encoding for the 16s rRNA region of Helicobacter spp. A relationship with gastric lesions was not found. The present study suggests that there is no direct relationship between helicobacteriosis and EGUS or EGGD, and that the infecting species of Helicobacter spp. in the horse stomach do not correspond to those with zoonotic potential. Helicobacter spp. DNA detection in equine tartar sets the foundations for conducting new epidemiologic studies about this condition, since this is the first report on the issue.