Introduction
Systemic lupus erythematosus (SLE) is a typical chronic autoimmune disease with a wide range of clinical manifesta tions and is characterized by the production of autoantibodies against nuclear self-antigens.1,2 Although various autoanti bodies production may be observed in SLE individuals, the antinuclear antibody (ANA) (3 is commonly used as a serological marker to diagnose SLE. The etiopathogenesis of SLE is incom pletely known. Nonetheless, genetic factors seem to play a pivotal part in disease susceptibility.
MicroRNAs (miRNAs) are highly conserved endogenous non-coding small RNA molecules and regulate the expres sion of protein-coding genes by either translational repression or messenger RNA degradation. (4 Among these, miR-146a and miR-499, are two widely reported genes that significantly con tribute to autoimmune diseases through their functional roles in modulating both innate and adaptive immune responses. (5-8 Therefore, both miRNAs could potentially responsible to the development and progression of SLE.
Interleukin (IL)-1 receptor-associated kinase (IRAK1) and tumour necrosis factor (TNF) receptor-associated factor 6 (TRAF6) have been reported as targets of miRNA-146a, (9 whereas miRNA-499 targets several factors including IL-17 receptor B (IL-17RB), (10,11 IL-23a, (12 IL-2R, (12 IL-6, (12,13 IL-2, (12 IL-18R, (12 and peptidyl arginine deiminase type 4 (PADI4), (14 in which all of these inflammatory mediators play crucial role in progressing the pathogenesis of autoimmune diseases, including SLE.
Sequence variations observed in miRNAs have been impli cated to influence the disease outcome. (3 This is possibly because single nucleotide polymorphisms (SNPs) within miR-NAs region may alter their expression and functions, (3,12 which then resulting in differential regulation of its tar get protein. Functional SNPs in both miR-146a and miR-499 genes have been evaluated in SLE patients from different populations. (12,15,16 However, the results remain inconclusive. Therefore, this study aims to assess the relationship between miR-146a, miR-499 and IRAK1 polymorphism and the risk of SLE.
Methods
Meta-analysis was performed in accordance to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. Records were identified through elec tronic databases dated up to September 2020 with search terms such as "miRNA-146a", "miRNA-499", "IRAK1" and "SLE". Studies were included on the basis of the following criteria: (1) aims to evaluate the association between gene polymorphism with predisposition to SLE; (2) conducted with a case-control design.
Meta-analysis for each gene polymorphism was performed for two or more studies. Genotypic frequency was tested for deviation from the Hardy-Weinberg equilibrium (HWE) in the control subjects. The association between gene polymorphism with predisposition to SLE was calculated by pooled odds ratio (OR) and 95% confidence interval (CI). The Z test was used to evaluate the significance of the pooled effect size. Study heterogeneity was evaluated using Q test and I2 statis tic. A significant Q-statistic (p<0.10) indicated heterogeneity across studies, with substantial heterogeneity indicated by an I2 value over 50%. The fixed-effect model (FEM) was used in the absence of heterogeneity, while the random-effect model (REM) was implemented if heterogeneity was present. A funnel plot and Begg's test were used to investigate the publication bias if the pooled effect size consisted of 10 or more studies. The value of 0.05 was indicative of the statistical significance. The Newcastle-Ottawa scale (NOS) was used to assess the study quality, in which a score ≥ 7 is considered a good study. (17-34
Results
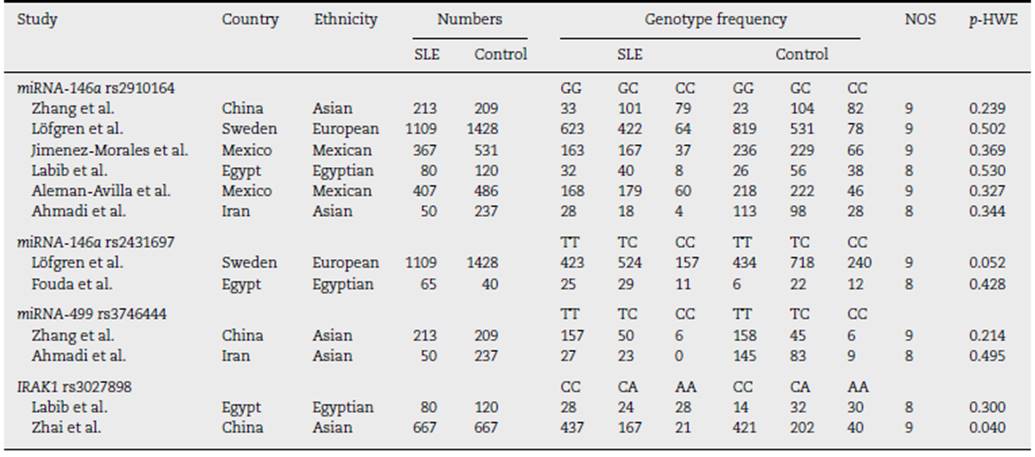
A total of 37 articles were screened, among which 27 were reviewed. Sixteen studies were excluded due to their absent relation to miRNA-146a, miRNA-499, and IRAK1 polymorphisms or the inability to extract the data. Eight studies were then included in this meta-analysis3,5,12,15,16,35-37 (Fig. 1). A total of 5237 (SLE: 2226, control 3011), 2642 (SLE: 1174, control: 1468), 709 (SLE: 263; control: 446), and 1534 (SLE: 747; control: 787) subjects for miRNA-146a rs2910164, miRNA-146a rs2431697, miRNA-499 rs3746444, and IRAK1 rs3027898 polymorphisms, respectively, were further analyzed. All studies complied with the HWE (Table 1). Details of the retrieved studies are depicted in Table 1.
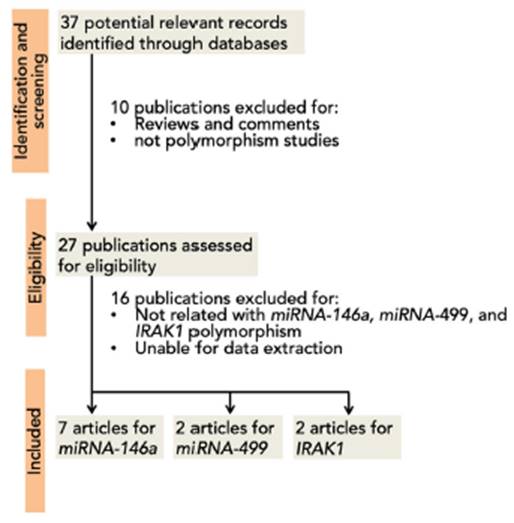
Fig. 1 PRISMA flow diagram of study selection process for the association between miR-146a, miR-499 and IRAK1 polymorphism and the risk of SLE.
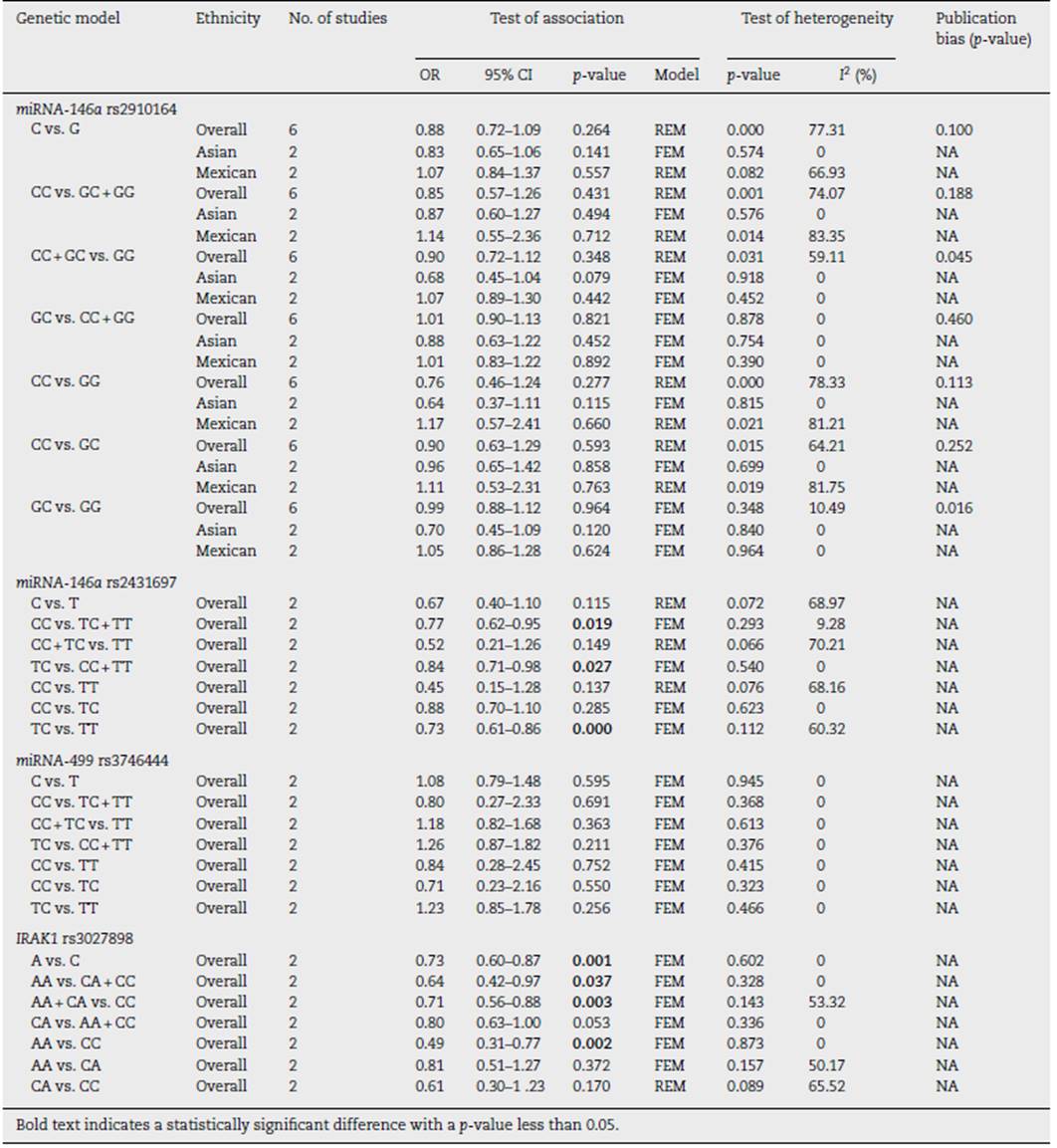
Pooled results on the associations between miRNA-146a, miRNA-499, and IRAK1 polymorphisms with SLE are shown in Table 2. We failed to show any significant association between miRNA-146a rs2910164 and miRNA-499 rs3746444 in any inheritance models (Table 2). However, we found that patients harbouring C allele of miRNA-146a rs2431697 were significantly lowered SLE predisposition (CC vs. TC + TT, OR = 0.77,95% CI = 0.62-0.95, p = 0.019; TC vs. CC + TT, OR = 0.84, 95% CI = 0.71-0.98, p = 0.027; and TC vs. TT, OR = 0.73, 95% CI = 0.61-0.86, p = 0.000, Table 2). In addition, IRAK1, target of miRNA-146a, was also examined. Patients with the A allele and AA genotype of rs3027898 were significantly decreased SLE susceptibility compare to the C allele and CC genotype, respec tively (A vs. C, OR = 0.73, 95% CI = 0.60-0.87, p = 0.001; AA vs. CA + CC, OR = 0.64, 95% CI = 0.42-0.97, p = 0.037; AA + CA vs. CC, OR = 0.71,95% CI = 0.56-0.88, p = 0.003, and AA vs. CC, OR = 0.49, 95% CI = 0.31-0.77, p = 0.002, Table 2). Whereas no significant difference between the AC and AA genotype (Table 2). No evi dence of publication bias was observed in any models in each gene polymorphisms (Table 2). Because we failed to show any association between miRNA-499 with SLE risk, genetic poly morphism of miRNA-499 target was not further evaluated.
Discussion
Our current study addresses the possible association between the miR-146a, miRNA-499, and IRAK1 polymorphism and its susceptibility to SLE. We found that the individuals carrying the C and A allele of rs2431697 and rs3027898, respectively, displayed protection against the occurrence of SLE, thereby implying that both miR-146a and its target protein, IRAK1, are crucial in the pathogenesis of SLE. Similar to previously reported studies38-41; our updated meta-analysis strengthens the notion that the miR-146a rs2910164 was not responsible for SLE predisposition.
Functional analysis revealed that numerous SNPs within miR-146a have been shown to affect the expression level of mature miR-146a. (16 For example, the T allele of rs2431697 decreases matures miR-146a expression. (16 Downregulation of human-miR-146a is associated to the manifestations of SLE by deregulating the activation of the interferon (IFN) pathway. (42,43 This may be due to several reason such as the decreasing expression of miR-146a and subsequent upregulation of IRAK1 and nuclear factor-KB (NF-KB) that may stimulate transcription of several genes-related inflamma tory responses. (44 Hence, it suggests the presence of inverse relationship between miR-146a expression level and SLE. Moreover, a study also identified that the biological axis between miR-146a and TRAF6 is closely linked to the SLE progression. (45 Additionally, mutant or alternative type (T allele) carriers of rs2431697 evidently increased the coronary artery disease risk, but not the severity of the disease. (46 Compiling to our findings, this implies that the rs2431697 is important in determining SLE susceptibility. Further studies investigating the effect of rs2431697 and SLE progression are required.
Although functional analysis of rs3027898 in IRAKI is cur rently unavailable, (47 it is possible to speculate that the C allele may contribute to the upregulation of IRAK1. Indeed, hyperactivation of IRAK1 in CD4+ T cells was observed in SLE individuals.48 While we failed to show any association between rs2910164 with SLE predisposition, other polymor phism such as rs57095329 in miR-146a is correlated with SLE. (40,39 Luo et al. (43 have successfully demonstrated the atten uation of miR-146 expression is partly influenced by low binding capability to Ets-1 due to conformational changes in the promoter region of rs57095329.
Association between miRNA-499 polymorphism with SLE was also evaluated, but no association was observed. Interest ingly, however, patients harbouring the T allele of rs3746444 had low RA risk. (39 Additionally, Lu et al. (49 demonstrated that rs3746444 polymorphism is closely linked to RA susceptibility in Mediterranean populations. Nonetheless, further studies are still required to determine whether the rs3746444 polymorphisms contribute to SLE susceptibility in different ethnic groups.
The present study has some limitations. Firstly, some of the pooled studies were generated from the limited num ber of included studies, thus the result may not be precise. Secondly, methodological variation of SNP identification and the opposite findings between studies may influence the results obtained in this particular study. Thirdly, because SLE is a complex disease, other factors, including, genetics, hor monal, environmental, and immune response/status should be closely examined.
In conclusion, our study demonstrates the association between miRNA-146a and IRAK1 polymorphisms with SLE risk. Further investigations evaluating functional analysis of miRNA-146a and IRAK1 in individuals with SLE and controls are necessary to clarify the role of the microRNA and its target gene in the etiology of SLE.