Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista MVZ Córdoba
Print version ISSN 0122-0268
Rev.MVZ Cordoba vol.18 no.3 Córdoba Sept./Dec. 2013
ORIGINAL
Genetic diversity of Prochilodus lineatus stocks using in the stocking program of Tietê River, Brazil
Diversidad genética de lotes de Prochilodus lineatus utilizados en el programa de repoblamiento del río Tietê, Brasil
Nelson Lopera-Barrero,1* Ph.D, Jayme Povh,2 Ph.D, Darci C. Fornari,3 M.Sc, Maria Rodriguez-Rodriguez,3 M.Sc, Silvio C. Alves dos Santos,4 Biol, Ricardo Ribeiro,3 Ph.D.
1Universidade Estadual de Londrina - Departamento de Zootecnia, Rod. Cendo Garcia Cid, Pr 445, km 380, CEP 86057-970 - Londrina, PR - Brazil.
2Universidade Federal de Mato Grosso do Sul - Faculdade de Medicina Veterinária e Zootecnia, Av. Senador Filinto Muller, no. 2443, CEP 79070-460 - Campo Grande, MS - Brazil.
3Universidade Estadual de Maringá, Departamento de Zootecnia, Núcleo de Pesquisa PeixeGen, Avenida Colombo, n° 5.790, CEP 87020-900, Maringá, PR, Brazil.
4Hidrelétrica AES Tietê - AES/Promissão, Rod. BR 153, km 139 - Promissão, SP - Brazil.
*Corresponding: nelson.peixegen@gmail.com
Recibido: Febrero de 2012; Aceptado: Marzo de 2013.
ABSTRACT
Objective. Assess the genetic diversity in four brood stocks and one juvenile stock of curimba Prochilodus lineatus in a Hydropower plant in São Paulo - Brazil, using the Tietê River stocking program. Materials and methods. Five RAPD primers were used to amplify the extracted DNA from 150 fin-clip samples. Results. Fifty-nine fragments were polymorphic, 52 had frequencies with significant differences (p<0.05), 45 had low frequencies, 54 were excluded, and two were fixed fragments. High values for polymorphic fragments (71.19% to 91.53%) and Shannon index (0.327 to 0.428) were observed. The genetic divergence values within each stock were greater than 50%. Most of the genetic variation was found within the groups through the AMOVA analysis, which was confirmed by the results of the identity and genetic distance. High ancestry levels (FST) among the groups value indicated high and moderate genetic differentiation. The estimates of number of migrants by generation (Nm) indicated low levels of gene flow. High and moderate genetic divergence between groups (0.58 to 0.83) was observed. Conclusions. The results indicate high variability within the stocks, and genetic differentiation among them. The fish stocks analyzed represent a large genetic base that will allow the fish technicians to release juveniles without genetic risks to wild populations present in the river. These genetic procedures may be used as models for other migratory species, including those threatened by extinction..
Key words: Fish, genetic diversity, RAPD (Source: DeCS).
RESUMEN
Objetivo. Evaluar la diversidad genética de cuatro lotes de reproductores y un lote de juveniles de curimba Prochilodus lineatus en una hidroeléctrica en São Paulo - Brasil, utilizados en el programa de repoblamiento del río Tietê. Materiales y métodos. Cinco primers RAPD se utilizaron para amplificar el ADN extraído de 150 muestras de aleta. Resultados. Cincuenta y nueve fragmentos fueron polimórficos, 52 tuvieron frecuencias con diferencias significativas (p<0.05), 45 tuvieron bajas frecuencias, 54 fueron excluidos, y dos fueron fragmentos limitantes. Se observaron altos valores de fragmentos polimórficos (71.19% a 91.53%) y de índice de Shannon (0.327 a 0.428). Los valores de divergencia genética dentro de cada lote fueron mayores al 50%. La mayor variación genética se encontró dentro de los grupos a través del análisis de AMOVA, el cual fue confirmado con los resultados de identidad y distancia genética. Los altos niveles de descendencia (FST) entre los grupos indicaron una alta y moderada diferenciación genética. Los cálculos del número de emigrantes por generación (Nm) indicaron bajos niveles de flujo génico. Se observó alta y moderada divergencia genética entre los grupos (0.58 a 0.83). Conclusiones. Los resultados indicaron una alta variabilidad dentro de los lotes y baja diferenciación entre ellos. Los lotes de peces analizados representan una amplia base genética que permitirá a los productores liberar juveniles sin riesgos genéticos para las poblaciones naturales presentes en el río. Estos procedimientos genéticos pueden ser utilizados como modelos para otras especies migratorias, inclusive en aquellas en peligro de extinción.
Palabras clave: Diversidad genética, pez, RAPD (Fuente: DeCS).
INTRODUCCIÓN
The Tietê is an important river in Brazil, located in the State of São Paulo. This River runs through the metropolitan area of São Paulo City and continues through 1.136 km up to the Paraná River, on the border with Mato Grosso do Sul. Along the way, several dams have been installed. These dams favor local and regional economic development, but they also bring serious and irreversible alterations of the natural hydrologic regime of rivers; affecting habitat quality, the dynamics of the biota and aquatic fauna, including fish (1). Recently, there has been a decline in the population of various wild fish like the Prochilodus lineatus (curimba) in this River.
P. lineatus (Prochilodontidae, Characiformes) is a migratory species of great economic importance for both fisheries and aquaculture, found throughout the Jacuí, Paraíba do Sul, Paraná, Paraguay and Uruguay River basins in South America (2). According to the Brazilian Ministry of Fisheries and Aquaculture (MPA), in 2009, 4.469 ton of Prochilodus were produced (3). The main factors responsible for the production decline in the Tietê River, have been the intense deforestation of the river banks, excessive fishing of juveniles, water pollution, introduction of exotic fish and hydropower dams (4-6). The Tietê River has more than 10 hydropower plants. These dams have broken migratory paths, interfered with the life cycle of numerous aquatic organisms, and produced major changes in ecosystems (7).
In dams, the passage ladders have been built to allow an alternative route of the natural flow of the migratory species. However, these ladders have been criticized for their unidirectional behavior (8), for their selectivity of the fish and their inefficiency in allowing downstream return of adults, eggs, and larvae (9). Another decision that has promoted the preservation of fish fauna is the regional development of stocking programs (10).
Although stocking programs have been used as a conservation method, risks associated with reducing the population size, increasing ecological competition between wild and captive-bred individuals, impact the ecosystem and reduce the genetic diversity of wild stocks; which are questionable (11,12).
In this context, the genetic variation of broodstocks used in these programs must be monitored to increase the preservation of wild fishing resources in stocking programs. With this purpose, the RAPD molecular marker (Random Amplified Polymorphic DNA) has been applied to estimate genetic diversity in broodstocks (13-15).
The objective of the present study was to evaluate the genetic diversity of Prochilodus lineatus stocks used in the stocking program of Tietê River, Brazil, by using RAPD markers.
MATERIAL AND METHODS
Collection of biological samples. The experiment took place between May 2010 and February 2011. We collected 120 fin-clip samples from four captive stocks of Prochilodus lineatus reared at the AES Hydropower plant (21°29'98"N and 49°78'40”W), São Paulo, Brazil. These stocks were kept in captivity for six years, and are important because they have been enhancing the fish stocking program in the Rivers of São Paulo. Were also collected 30 samples of juveniles (90 days old) to assess the genetic variability of individuals being released in the stocking process. The origin of stocks:
Broodstock A (BA): second generation (F1) bred from wild populations collected from the Tietê River, São Paulo.
Broodstock B (BB): first generation (F0) from wild populations collected in the Pardo River, São Paulo.
Broodstock C (BC) and D (BD): are of unknown origin. However, it is known that the BC has the first generation of individuals in captivity (F0), and BD has second-generation (F1) individuals in captivity.
Juvenile stock (JS): unknown origin.
Molecular methods. The DNA was isolated from the 0.5 cm2 fin-clips, and extraction was based on the previously described methods (16). The samples were treated with 550 ml lise buffer (50 mM Tris-HCl, 50 mM EDTA, 100 mM NaCl, and 1%(w/v) SDS), 7 ml proteinase K (200 µg/ml) per sample, and incubated overnight at 50°C. Then, 600 µl 5 M NaCl was added to each sample before being centrifuged for 10 min at 12000 rpm. The aqueous layer was removed carefully into new micro tubes where the DNA was precipitated with 700 µl of freezing ethanol and incubated later at -20°C for 2 h. The DNA samples were centrifuged again, washed with 700 µl 70% (v/v) ethanol, re-suspended in TE buffer (10 mM Tris and 1 mM EDTA), and treated with 6 ml RNAse (30 µg/ml) at 38°C for 40 min. The DNA was quantified in the Shimadzu spectrophotometer (UV-1601, USA) with absorbance at 260 nm. The samples were diluted to the concentration of 10 ng/µl. The DNA quality was checked using agarose gel electrophoresis (1% w/v) buffered with TBE 1X (500 mM Tris-HC1, 60 mM boric acid and 83 mM EDTA) at 70 volts for 1 h. Thereafter, the gels were photographed using the L-PIX Image Transiluminator software 1.3 - Loccus Biotechnology - Molecular Image, USA (Figure 1).
The genomic DNA was amplified in the reaction volume of 15 ml using buffer Tris-KCl 1X (Tris-HCl 20 mM pH 8.4 and KCl 50 mM), 2.5 mM MgCl2, 0.46 mM primer (oligonucleotides), 0.2 mM from every dNTPs, one unit of Platinum Taq DNA Polymerase and 10 ng of DNA. The RAPD reactions were amplified in the “Eppendorf Mastercycler® Gradient” thermocycler, programmed to 40 cycles, an initial step of denaturation at 94°C for 4 min, and a final step of extension at 72°C for 5 min. Every cycle consisted of 1 min at 94oC, 90 s at 40°C and 2 min at 72°C.
We evaluated 60 primers from the Operon Kit (Operon Technologies Inc. in Alameda, California, USA). Different stocks were evaluated for five selected primers. The product from the amplification was separated in agarose gel at 1.4% (w/v). We used 15 ml of the amplified product and 2 ml of the sampling buffer (40% (w/v) sucrose and 0.25% (w/v) bromophenol blue) in horizontal electrophoresis. The electrophoresis was carried out at 70 V for 4 h (3 V/cm) using the TBE 1X buffer. Every reaction had a negative control (N) where all the previous components but DNA were added to the solution. A bath with ethidium bromide at 0.5 mg/ml for 30 min was used to reveal the gel. Thereafter, the gels were photographed using the L-PIX Image Transiluminator software 1.3.
RAPD statistical analysis. The size of the fragments was determined by comparison with a 100 pb DNA Ladder (Invitrogen®, USA). The presence or absence of identical molecular-sized fragments was used to construct a similarity matrix based on the calculation of the Jaccard similarity coefficient, codifying 1 if a fragment was present and 0 if absent.
Based on this matrix, the genetic divergence was estimated by the Mantel test, using the Monte Carlo method, by Mantel-Struct program (17). The percentage of polymorphic fragments and the Shannon index were obtained from the POPGENE software version 1.31 (18). The software TFPGA 1.3 (19) was used to estimate the identity and genetic distance among the groups. The frequency of fragments was estimated by the exact test.
The ARLEQUIN 3.0 software (20) was used to determine the genetic differentiation using the estimates of the ancestry coefficient (FST), the number of migrants per generation (Nm) and the molecular analysis of variance - AMOVA. The molecular variance between the groups was evaluated after combining all of them in the following groups: BAxBB, BAxBC, BAxBD, BAxJS, BBxBC, BBxBD, BBxJS, BCxBD, BCxJS and BDxJS. The significance of these tests was determined by the random permutation method using from 1.000 to 10.000 permutations. The significance of the FST was tested by the X2 [c2=2n Fst (k-1); GL = (k-1) (s-1)] after (21) in which n is the number of individuals in the two groups, k is the number of alleles and s is the number of groups. The magnitude of the genetic differentiation between these groups was based on the Wright definition (22) as having little differentiation when FST is ranging between 0 and 0.05, moderate when FST is ranging between 0.051 and 0.15, high when the FST is ranging between 0.151 and 0.25 and very high when the FST is above 0.25.
RESULTS
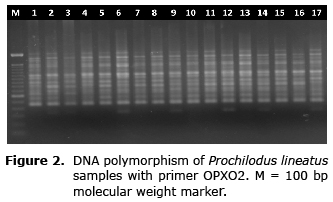
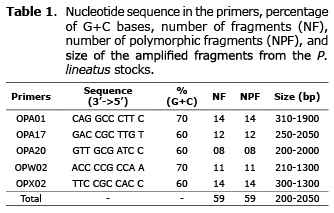
Fifty-nine amplified fragments were polymorphic (100%). The number of clear and reproducible fragments generated per primer ranged from eight (primer OPA20) to 14 (primer OPA01 and OPX02) (Figure 2). The biggest fragment (2050bp) was obtained from the primer OPA17 and the smallest (200bp) from the primer OPA20 (Table 1).
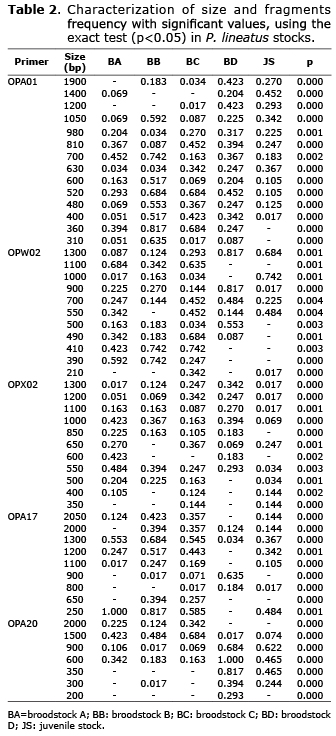
Fifty-two in 59 fragments had significant differences in their frequencies (p<0.05). A total of 45 fragments with low frequency (lower than 0.100) were found in the five stocks (BA=11; BB=7; BC=11; BD=5; JS=11). Fifty-four were excluded fragments - frequency of 0.000 (BA=11; BB=11; BC=5; BD=14; JS=13), and two were fixed fragments - frequency of 1.000 (BA=1; BD=1). Exclusive fragments were not found (Table 2).
The percentage of polymorphic fragments FP (BA=77.97, BB=89.83, BC=91.53, BD=71.19 and JS=77.97%) and the Shannon index - SI (BA=0.352, BB=0.411, BC=0.428, BD=0.369 and JS=0.327) were higher in all the stocks. The genetic divergence (GDIV) values within each stock were greater than 50% (BA=0.62; BB=0.53; BC=0.56; BD=0.53; JS=0.67), showing also a high variability.
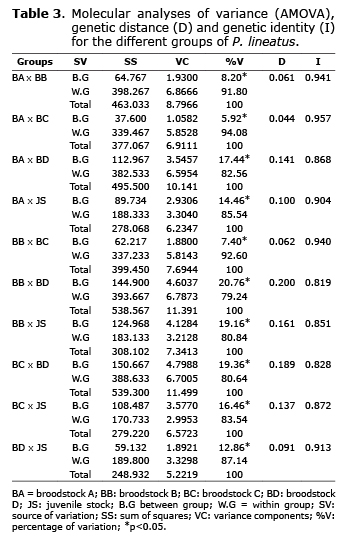
According to the analysis of molecular variance (AMOVA), most of the genetic variations occurs within every group of P. lineatus and not between them. These results were confirmed by the estimates of identity and genetic distance (Table 3).
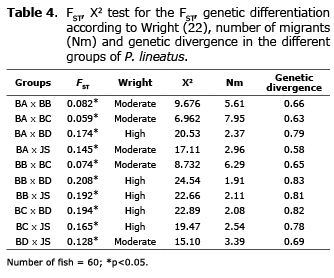
The FST showed high ancestry between groups. These results suggest a high and moderate genetic differentiation. Corroborating this hypothesis, the Nm was low in all the groups suggesting a low gene flow. High and moderate genetic divergence between groups was also observed (Table 4).
The FST showed high ancestry between groups. These results suggest a high and moderate genetic differentiation. Corroborating this hypothesis, the Nm was low in all the groups suggesting a low gene flow. High and moderate genetic divergence between groups was also observed (Table 4).
DISCUSSION
The RAPD analysis was appropriate to evaluate the genetic diversity within and between P. lineatus stocks.
The main problems towards the dominant performance of this technique (23) were overcome through the amplification tests and standardization of samples carried out in conjunction with the negative controls in the different amplifications.
Forty-five fragments with low frequency and fifty-five excluded fragments were found in the five stocks (Table 2). According to Jacometo et al (24), the presence of low frequencies and absence of these fragments within each stock may be explained by the bottleneck effect or founder effect. The bottleneck effect is the reduction of an existing population to a small number of individuals. These decrease causes the loss of genetic and allele variability (25). The founder effect is the formation of stocks with few individuals that normally do not represent the genetics of natural populations. This effect may result from the presence of low frequency or absent alleles (24). Two fragments in BA and BD were fixed. This result showed that, despite the presence of fragments with low frequency or absent, there is was a high genetic variability within both stocks.
The results for genetic variability and divergence, calculated by FP, SI and GDIV exhibited high genetic variability in four broodstocks. These results showed that broodstocks were formed from a large number of individuals with high genetic variation, which allowed the maintenance of variability. The gene pool of fish broodstocks in aquaculture is sometimes characterized by reduced diversity (26). inadequate reproductive management as well as a small number of effective contributing parents (Ne) (27), inappropriate selection of parents, founder effect, misuse of the reproduction period (15) and the breeding system used in mating (28) are also some constraints capable of inducing significant loss in the genetic variability.
Juvenile stock also exhibited high genetic variability. Reproductive management is highly important to preserve the genetic diversity of juveniles, which will participate in the stocking programs. An inadequate reproductive management may cause less survival of juveniles and consequently, the wild populations will be permanently affected. Similar results were observed in Brycon orbignyanus offspring used in stocking programs; where the genetic variability has been preserved by an appropriate reproductive management (14). In contrast, Lopez et al (29) observed a decrease in genetic variability in P. lineatus offspring used in stocking programs (broodstock: %FP=85.71, offspring: %FP=79.37), possibly due to inadequate reproductive management.
The AMOVA analysis, genetic distance (GDIS) and identity (GID) showed similarity between BA, BB and BC, which reflects a common origin or individual exchange between broodstocks. The similarity between BA and BB is surprising, since the two groups have different origin. Founder effect or no intentional exchange between them during reproductive management may have contributed to this similarity. Unfortunately, the management decisions that were made during the reproductive season are unknown. Analyzing two juvenile stocks of P. lineatus, Lopera-Barrero et al (30) did not find genetic differentiation (GDIS: 0034; GID: 0967) due to the founder effect and reproductive management.
The FST and genetic divergence showed high ancestry and divergence between groups, which suggests a high and moderate genetic differentiation. Corroborating this hypothesis, the Nm was low in all the groups suggesting a low gene flow (Table 4). The FST (0.128), Nm (3.39) and genetic divergence - GDIV (0.69) values suggest that BD is the parent origin of the juvenile stock, which was corroborated by the genetic distance (0.091). However, it is not possible to dismiss the paternity of BA, which also shows similarity values (FST: 0.145, Nm: 2.96, GDIV: 0.58, GDIS: 0.100).
BA x BC (FST: 0.059, Nm: 7.95, GDIV: 0.63, GDIS: 0.044) and BB x BC (FST: 0.074, Nm: 6.29, GDIV: 0.65, GDIS: 0.062), also revealed little genetic differentiation. Due to the absence of management information, it is not possible to state that they have a common origin or individual exchange between the broodstocks. Moreover, BC showed larger genetic variability than BA and BB (%FP: 91.53, IS: 0.428).
Based on these results, it is suggested that the reproductive and genetic management of four P. lineatus broodstocks analyzed in this study, should be considered as genetically different stocks. This suggestion should be applied especially when stocks are used in repopulating or breeding programs. This way, the first step in the establishing hatcheries and stocking programs, is to investigate the genetic variability of broodstocks (30). The correct selection of the individuals to be used in the formation of broodstock is a crucial step (27), and their genetic evaluation can offer important bases for formulating reproductive management strategies. These strategies will allow safe exchange of broodstocks among fish farms to break cycles of endogamy which are common in controlled environments (25).
Conversely, the decline in the genetic variability may induce adaptability and survivorship problems for these offspring, and negatively influences on the ecosystem (28). In a worst-case scenario, species can go extinct (4). Thus, all the fish released during stocking programs should represent wild populations (5), and both must undergo genetic analysis.
Genetic monitoring using molecular markers such as the RAPD on the wild populations, broodstocks and progenies that have participation in stocking programs is fundamental to avoid declines in the genetic variability, avoiding effects on wild populations, and allowing for insights into preservation, management and reproduction of the P. lineatus.
In summary, the current results indicated high variability within the stocks and genetic differentiation among them. The fish stocks analyzed represent a large genetic base that will allow fish technicians to release juveniles without genetic risks to wild populations. Few studies have been conducted in this species, genetically evaluating broodstocks and offspring (29,30). Therefore, the results are important, especially when are used in stocking, breeding or conservation programs.
Acknowledgements
The authors thank to the AES Hydropower plant for providing the funds and samples for this study, the undergraduate students and the technicians for the important contributions to the success of this experiment.
REFERENCES
1. Agostinho AA, Pelicice FM, Gomes LC. Dams and the fish fauna of the Neotropical region: impacts and management related to diversity and fisheries. Braz J Biol 2008; 68:1119-1132. [ Links ]
2. Garcez R, Calcagnotto D, Almeida-Toledo LF. Population structure of the migratory fish Prochilodus lineatus (Characiformes) from rio Grande basin (Brazil), an area fragmented by dams. Aquat Conserv 2011; 21:268-275. [ Links ]
3. Lopera-Barrero NM, Ribeiro RP, Povh JA, Vargas Mendez LD, Poveda-Parra AR. Produção de organismos aquáticos: uma visão geral no Brasil e no mundo. Brasil: Rígel & Livros; 2011. [ Links ]
4. Agostinho AA, Thomaz SM, Gomes LC. Conservation of the biodiversity of Brazil's inland waters. Conserv Biol 2005; 19:646-652. [ Links ]
5. Lopera-Barrero NM. Conservation of Brycon orbignyanus natural populations and stocks for their reproductive, genetic, environmental sustainability: A model for species threatened with extinction. Cien Inv Agr 2009; 36:191-208. [ Links ]
6. Viana AP, Lucena Frédou F, Frédou T, Torres MF, Bordalo AO. Fish fauna as an indicator of environmental quality in an urbanized region of the Amazon estuary. J Fish Biol 2010; 76:467-486. [ Links ]
7. Carolsfed J, Harvey B, Baer A, Ross C eds. Migratory fishes of South America: Biology, Social Importance and Conservation Status. Victoria: World Fisheries Trust; 2003. [ Links ]
8. Britto SGC, Sirol RN. Transposição de peixes como forma de manejo: as escadas do Complexo Canoas, médio rio Paranapanema, bacia do Alto Rio Paraná. En: Nogueira MG, Henry R, Jorcin A eds. Ecologia de Reservatórios: Impactos Potenciais, Ações de Manejo e Sistemas em Cascata. São Carlos: Rima; 2006. [ Links ]
9. Agostinho AA, Marques EE, Agostinho CS, Almeida DA, Oliveira RJ, Melo JRB. Fish ladder of Lajeado Dam: migrations on one-way routes?. Neotrop Ichthyol 2007a; 5:121-130. [ Links ]
10. Povh JA, Ribeiro RP, Lopera-Barrero NM, Jacometo CB, Vargas L, Gomes PC et al. Microsatellite analysis of pacu broodstocks used in the stocking program of Paranapanema River, Brazil. Sci Agric 2011; 68:308-313. [ Links ]
[vcitat][text-ref]10. Massone AR, Martín AA, Ibargoyen GS, Ofek I, Stavri H. Immunohistochemical methods for the visualization of Mycobacterium paratuberculosis in bovine tissues. J vet med B 1990; 37:251-25311. Araki H, Schmid C. Is hatchery stocking a help or harm? Evidence, limitations and future directions in ecological and genetic surveys. Aquaculture 2010; 308:2-11. [ Links ]
12. Kitada S, Shishidou H, Sugaya T, Kitakado T, Hamasaki K, Kishino H. Genetic effect of long-term stock enhancement programs. Aquaculture 2009; 290:69-79. [ Links ]
13. Gomes PC, Ribeiro RP, Lopera-Barrero NM, Povh JA, Vargas L, Sirol RN. Genetic diversity of three stocks of piapara (Leporinus elongatus) using RAPD. Acta Sci Anim Sci 2008; 30:241-247. [ Links ]
14. Lopera-Barrero NM, Ribeiro RP, Sirol RN, Povh JA, Gomes PC, Streit Jr DP et al. Genetic characterization of Brycon orbignyanus stocks used in stocking programs. Rev MVZ Córdoba 2008; 13:1110-1119. [ Links ]
15. Povh JA, Ribeiro RP, Lopera-Barrero NM, Gomes PC, Blanck DV, Vargas L et al. Monitoring of the genetic variability of pacu, Piaractus mesopotamicus, of the stock enhancement program of the Paranapanema River. Arq Bras Med Vet Zootec 2009; 61:1191-1195. [ Links ]
16. Lopera-Barrero NM, Povh JA, Ribeiro RP, Gomes PC, Jacometo CB, Lopes TS. Comparison of DNA extraction protocols of fish fin and larvae samples: modified salt (NaCl) extraction. Cien Inv Agr 2008; 35:65-74. [ Links ]
17. Miller M. Mantel-Struct: a program for the detection of population structure via mantel tests. J Hered 1999; 90:258-259. [ Links ]
18. Yeh FC, Boyle TYZ, Xiyan JM. POPGENE Version 131: Microsoft Window-based freeware for population genetic analysis. Alberta: University of Alberta and Center for International Forestry Research; 1999. [ Links ]
19. Miller MP. Tools of population genetic analysis (TFPGA) 1.3: a Windows program for the analysis of allozyme and molecular population genetic data. Utah, Utah State University; 1997. [ Links ]
20. Excoffier L, Laval G, Schneider S. Arlequin Ver. 3.1: an integrated software package for population genetics data analysis. Evol Bioinform Online 2005; 1:47-50. [ Links ]
21. Workman PL, Niswander JD. Population studies on southwestern Indian tribes. II. Local genetic differentiation in the Papago. Am J Hum Genet 1970; 22:24-49. [ Links ]
22. Wright S. Evolution and genetics of populations. Chicago: University of Chicago Press; 1978. [ Links ]
23. Liu ZJ, Cordes JF. DNA marker technologies and their applications in aquaculture genetics. Aquaculture 2004; 238:1-37. [ Links ]
24. Jacometo CB, Lopera-Barrero NM, Rodriguez-Rodriguez M del P, Gomes PC, Povh JA, Streit Jr DP et al. Genetic variability of tambaqui (Teleostei: Characidae) from different regions of Brazil. Pesq agropec bras 2010; 45:481-487. [ Links ]
25. Moreira AA, Hilsdorf AWS, Silva JV, Souza VR. Genetic variability of two Nile tilapia strains by microsatellite markers. Pesq Agropec Bras 2007; 42:521-526. [ Links ]
26. Fopp-Bayat D, Jankun M, Kuzminski H. Genetic characterization of Polish culture brook trout, Salvelinus fontinalis (Mitchill), based on microsatellite DNA analysis. Arch Pol Fish 2010; 18:93-99. [ Links ]
27. Borrel YJ, Carleos CE, Asturiano JF, Bernardo D, Vázquez E, Corral N et al. Use of microsatellites and a combinatorial optimization approach in the acquisition of gilthead seabream (Sparus aurata L.) broodstocks for hatcheries. Aquaculture 2007; 269:200-210. [ Links ]
28. Lopera-Barrero NM, Vargas L, Sirol RN, Ribeiro RP, Povh JA, Mangolin CA. Genetic characterization of Brycon orbignyanus using the semi-natural system. Arq Bras Med Vet Zootec 2010; 62:184-191. [ Links ]
29. Lopes TS, Ribeiro RP, Lopera-Barrero NM, Sirol RN, Povh JA, Gomes PC et al. Genetic characterization of curimba (Prochilodus lineatus) stocks used in stock enhancement programs. Rev Bras Saúde Prod An 2008; 9:652-661. [ Links ]
30. Lopera-Barrero NM, Ribeiro RP, Vargas L, Povh JA, Gomes PC, Mangolin CA et al. Genetic characterization of Prochilodus lineatus (Valenciennes, 1836) stocks using in stocking programs: importance for the ichthyofauna conservation and ecosystem. Biosci J 2008; 24:86-93. [ Links ]