Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Ciencia y Tecnología Agropecuaria
Print version ISSN 0122-8706
Corpoica cienc. tecnol. agropecu. vol.13 no.2 Mosquera June/Dec. 2012
MICROBIOLOGÍA DEL SUELO
1 Centro de Biotecnología y Bioindustria, Corporación Colombia de Investigación Agropecuaria (Corpoica). Mosquera (Colombia). rbonilla@corpoica.org.co
Fecha de recepción: 26-04-2012. Fecha de aceptación: 24-07-2012
ABSTRACT
For mass plant growth-promoting inoculant production, a high-yield culture medium is fundamental. Sequential application of statistical designs was used to optimize Azospirillum brasilense C16 biomass production. Six nutritional (glycerol, glutamate, mannitol, citric acid, yeast extract and K2HPO4 3H2O) and three mineral sources (MgSO4 7H2O, FeCl3 and NaCl) were evaluated using five statistical experiments Placket-Burman, factorial design, steepest ascent, response surface analysis, and mineral screening. The optimum medium composition (g L-1) was as follows: 28.33 glutamate, 2.92 yeast extract, 1.34 K2HPO4 3H2O, 0.5 MgSO4 7H2O and 0.02 FeCl3. After 24 hours of incubation, protein (32.04 mg) and dry biomass (1.51 g L-1) were 1.72 and 1.68 times higher than in conventional growth medium.
Key words: culture medium optimization, factorial designs, culture medium design, bioinoculant production
RESUMEN
Para la producción masiva de inoculantes basados en bacterias promotoras de crecimiento vegetal (PGPR), es fundamental un medio de cultivo de alto rendimiento. La aplicación secuenciada de diseños estadísticos fue usada para optimizar la producción de biomasa de Azospirillum brasilense C16, seis fuentes nutricionales (glicerol, glutamato, manitol, ácido cítrico, extracto de levadura y K2HPO4 3H2O) y tres fuentes minerales (MgSO4 7H2O, FeCl3 y NaCl) fueron evaluadas mediante cinco experimentos estadísticos - Placket-Burman, factorial fraccionado, diseño de paso ascendente, análisis de superficie de respuesta y screening mineral, para tal efecto. La composición optimizada del medio (g L-1) fue: 28,33 glutamato, 2,92 extracto de levadura, 1,34 K2HPO4 3H2O, 0,5 MgSO4 7H2O y 0,02 FeCl3, la cual luego de 24 h de incubación permitió producir una cantidad de proteína (32,04 mg) y biomasa seca (1,51 g L-1) del 1,72 y 1,68 veces más alta, respectivamente, en relación al medio de cultivo convencional.
Palabras clave: optimización de medio de cultivo, diseños factoriales, diseño de medio de cultivo, producción de bioinoculantes
INTRODUCTION
The Azospirillum genus has been widely studied in recent years. This microorganism has usually been associated with the rhizosphere of both cereals and grasses, being recognized as plant growth-promoting bacteria (PGPB) (Bashan and de-Bashan, 2010; Arzanesh et al., 2011; Bashan et al., 2011). Bacterial synthesis of growth-promoting substances -cytokinins, gibberellins, and indole- has demonstrated the ability to improve root development and enhance water and mineral uptake (Dobbelaere and Okon, 2007; Spaepen et al., 2007). These capabilities have made this PGPB suitable for application to corn, rice, and wheat crops (Díaz-Zorita and Fernández-Canigia, 2009; Hartmann and Bashan, 2009; Bashan et al., 2011).
The goal of PGPB inoculant design is their application by growers (Bashan et al., 2011). Inoculants are based on high numbers of viable cells in standardized formulations. Fo mass production of microorganisms -in flasks as well as bioreactors-, an optimized, defined, and yet cheap culture medium is necessary. Different strategies for optimization of culture media have been applied (Mendes et al., 2001; Liu et al., 2003; Wang et al., 2007), but as Ren (2008) described, very few papers have applied all experimental designs well. An alternative culture medium de novo design and its optimization require an integrative application of experimental designs. Screening of nutritional factors, optimization of a subset of components (steepest ascent analysis), and application of a response surface methodology are steps to obtain the maximum response (Liu et al., 2003; Ren et al., 2008).
Although Bashan et al. (2011) reported two new growth media based on substitution of nutritional sources, to our knowledge, no studies have focused on the de novo design of a culture medium for Azospirillum brasilense mass production. Hence, our goal was to establish the most suitable nutritional conditions to optimize the biomass yield for inoculant production based on the use of this PGPB, using a sequential statistical approach.
MATERIALS AND METHODS
Bacterial strain and culture conditions
Azospirillum brasilense C16 was provided by the Banco de Germoplasma de Microorganismos con Potencial Biofertilizante - Corpoica, Mosquera, Colombia. Strain C16 was isolated from the grass Panicum maximum Jacq. in a sylvopastoral system in Cesar, Colombia, and selected based on its potential to be used as an inoculant for grasses (Cárdenas et al., 2010). The strain was grownth and maintained on a DYGS culture medium (composition g L-1: 2.0 glucose; 2.0 malic acid; 2.0 yeast extract; 1.5 peptone; 0.5 K2HPO4 3H2O; 0.5 MgSO4 7H2O; 1.5 L glutamic acid; pH 6.0). The cultures were incubated for 48 h at 120 rpm and 28±2°C.
Experimental strategy
Sequential statistical designs were used to investigate the effect of nutritional sources on biomass production. First, a Plackett-Burman (P-B) design was used to identify the sources with positive effects on the response variable (Plackett and Burman, 1946). Applying a factorial design, we focused on the critical subset of positive effect sources, identifying positive interactions. Then, the optimal region was found using the steepest ascent method (Ren et al., 2008). Finally, the selected sources were optimized by employing a response surface analysis "Box-Behnkendesign" (Box and Behnken, 1960). Additionally, mineral screening was performed to identify C16 minor growth needs.
Experimental conditions
The experiments were carried out in 50 mL flasks at 28°C, 150 rpm, and incubation for 24 h standard conditions. The pH was not controlled during the time of the experiment;Â to let the microorganism select nutritional sources with the best physicochemical effect with the statistical model. Each flask was inoculated with 200 mL of a twice-washed bacterial suspension adjusted to DO600 = 0,500. The total protein content of washed cells was measured as a response variable as reported by Bradford (1976).
Biomass production trial
A comparative trial between both the alternative and DYGS media was carried out. Both media were compared by their protein (mg) and weight biomass (dry weight in g L-1) production under the conditions described above for the statistical designs.
Experimental design and statistical analysis
Each statistical design was tested in three replicates (blocks), where a single Erlenmeyer flask was set as the experimental unit. Statistical experimental designs used in this paper were generated and analyzed using the statistical packages: Statgraphics Centurion XV (version 15, Statgraphics, USA) and SPSS 19 (SPSS®, IBM).
RESULTS AND DISCUSSION
Screening stage: Plackett-Burman design
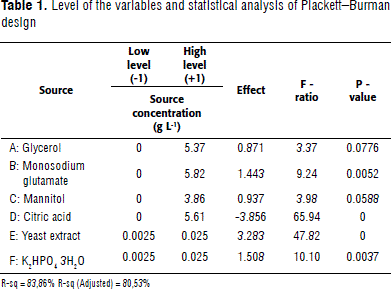
Due to the large number of media components assessed in this studywork, running experiments would cost time and money, so a P-B design was chosen to screen important sources. Six nutritional sources, including glycerol, monosodium glutamate, mannitol, citric acid, yeast extract, and K2HPO4 3H2O, were selected based on the metabolical characteristics previously described by Baldani et al. (2005). Two levels were established for each factor: low (-1) and high (+1) (Table 1). Levels for nutritional sources were set by adjusting:
High levels (+1) for C, N, and P were considered at the same concentrations as DYGS. The amount of total carbon was adjusted to 2.1 g L-1 carbon. When more than one C source was present, their amounts were equally divided, resulting in the same total carbon concentration. Yeast extract and monosodium glutamate were though just as nitrogen sources, ignoring additional minor nutritional contributions.
Low levels (-1) for glycerol, glutamate, mannitol, and citric acid were set to 0 g L-1. For yeast extract and K2HPO4 3H2O, low levels were set at 10% of the high level.
P-B design resolution is III, this means that the main effects are not confused. However, one or more than two-way interactions may be confounded; therefore, it must be assumed to be zero for main effects are meaningful. Results showed that media components having a positive effect were: monosodium glutamate, yeast extract, and K2HPO4 3H2O (P ≤ 0.05). Citric acid and glycerol were removed due to their negative and non-significant effects, respectively (P ≥ 0.05) (Table 2). Mannitol was kept despite its significance, in order to maintain two carbon sources mannitol and monosodium glutamate for the next design. Interestingly, we did not found a positive response when glycerol was assessed, in contrast to the results obtained by Bashan et al. (2010). We found that glutamate and yeast extract were notable available growth sources.
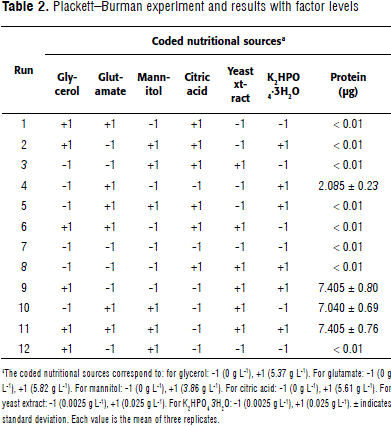
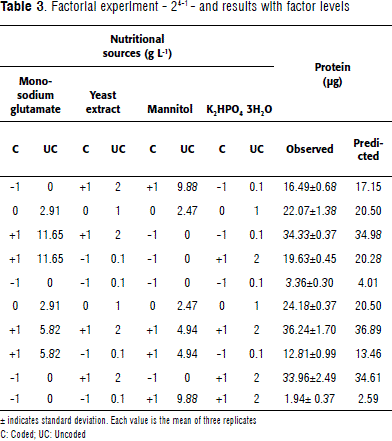
Primary optimal stage: factorial design
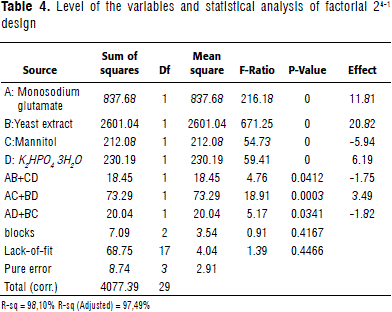
The resolution of the factorial design used was IV; meaning that both the main and two-way effects are not confounded. Concentrations of the selected sources: monosodium glutamate, mannitol, yeast extract, and K2HPO4 3H2O were evaluated with a 24-1 factorial design -in an effort to reposition the experimental region-seeking a more optimal region. For data analysis, the mid-level (for the center points) was coded as zero. Substrate high levels were doubled from the center point concentrations, and low levels were kept as in the screening design. The model could explain 98.09% of the variation (R-sq) present in the system. Observed and predicted protein values were close, showing the model's goodness of fit and the lack-of-fit value determined that the current model adequately represents the observed data (Table 3). Mannitol had a negative effect on protein production and therefore was removed (Table 4).
Confirmation stage: steepest ascent method
A necessary condition for a unique maximum is the presence of negative quadratic effects; if the optimal region for running the process has been identified, a response surface design is appropriated. The factorial design could explain second-order interactions but not quadratic effects. Therefore, additional experiments should be performed to seek the optimal region. Directional search methods -steepest ascent and ridge analysis- use the magnitude and sign of the linear effects to determine the direction toward a higher response. The steepest ascent path begins at the center of the current design space -optimal levels from previous design- and stretches well outside the design space. A sequence of equally spaced locations along the path is then selected which form a set of experiments (Ren et al., 2008).
Based on the optimal source concentrations determined from the factorial design, three nutritional sources -monosodium glutamate, yeast extract and K2HPO4 3H2O- were identified, selected, and applied in the steepest ascent method. A four step path was designed from the optimal nutrient levels obtained from the factorial design -named as D step-, and the concentrations were increased by twice, three-, and four-times, respectively. From the results, it was possible to identify that step 2D was the most suitable for biomass production (Table 5).

Optimization stage: Box-Behnken design
On the basis of step 2D, a response surface analysis was executed. Concentrations of the selected sources: glutamate, yeast extract, and K2HPO4 3H2O were adjusted up (+1) and down (-1) from the center point with a 50% variation (Table 6). The multiple determination coefficient (R-sq) indicated that the model could explain 86.19% of the variation present in the system (Table 7). A non-lineal expression was deduced from the obtained results, which explained the biomass production (Equation 1).
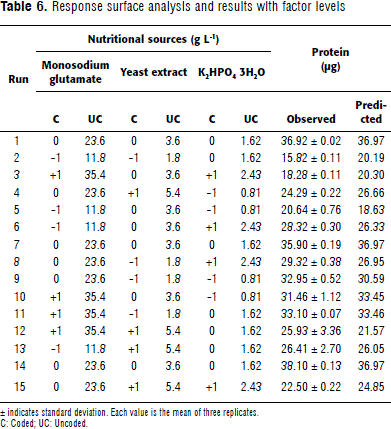
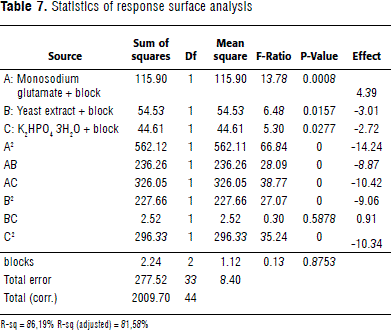
Where Y is mg of protein produced in 24 h; A, B and C are the values of monosodium glutamate, yeast extract and K2HPO4 3H2O, respectively.
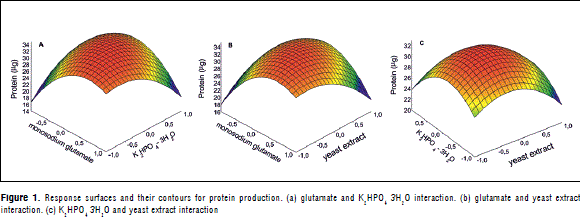
The predicted protein values were very close to the experimentally obtained ones, indicating the model's goodness of fit. The response surfaces and their contours determined each optimum variable level for maximum productivity (Figure 1A, B and C). The model predicted a maximum of 37.94 mg protein in 24 h, with the optimum concentrations of 28.33 g L-1 of monosodium glutamate; 2.92 g L-1 yeast extract, and 1.34 g L-1 K2HPO4 3H2O. Three second-order polynomial equations were found to explain protein production (Equations 2, 3, and 4).
Where Y is mg of protein produced in 24 h; A, B and C are the values of monosodium glutamate, yeast extract and K2HPO4 3H2O, respectively.
Selected sources for growth of A. brasilense C16 were glutamate, yeast extract, and dibasic potassium phosphate. Under aerobic conditions, glutamate is reported to allow both growth and nitrogen fixation in A. brasilense and A. halopraeferens (Baldani et al., 2005). Glutamate and yeast extract interaction may be based on: 1) yeast extract effect has been reported to decrease lag phase and stimulate vigorous growth (Peña et al., 1997; Bashan et al., 2011), and 2) glutamate is usually assimilated at a low rate (Baldani et al., 2005). Hence, yeast extract may be used as a main nutrient source allowing later expression of the glutamate metabolizing enzymes: glutamate dehydrogenase (GDH, glutamate-oxaloacetate-aminotransferase (GOT), and glutamate-pyruvate-aminotransferase (GTP). Interestingly, glutamate exhibited the most significant effect in all the statistical designs, being used as the main growth source by A. brasilense C16.
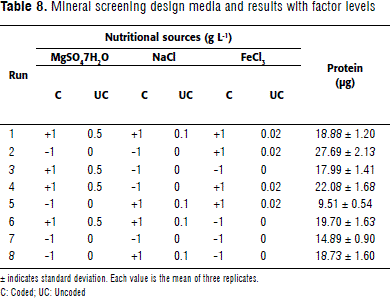
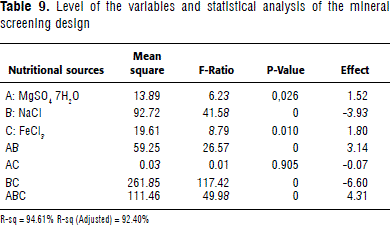
Mineral screening design
Based on optimal source concentrations for A. brasilense C16 -from response surface analysis-, a quick screening design for minerals was applied. Minor nutritional needs from MgSO4 7H2O, NaCl and FeCl3 salts were identified (Table 8). The Mg and Fe salts used exhibited a positive effect on bacterial growth, while NaCl had a negative one. Interestingly, divalent metals optimized biomass production, even taking into account that yeast extract might cover minor nutritional requirements (Table 9). Mg and Fe have been described as playing a role as protein cofactors and being associated with important metabolic responses, such as nitrogen fixation. So, the optimal source concentrations that maximized biomass production were: 28.33 g L-1 monosodium glutamate, 2.92 g L-1 yeast extract, 1.34 g L-1 K2HPO4 3H2O, 0.5 g L-1 MgSO47H2O, and 0.02 g L-1 FeCl3.
Biomass production trial
Our alternative media produced 1.71-times more protein than DYGS in 24h (Figure 2). Similarly, biomass production exhibited 1.68 times more production than DYGS (0.9 g L-1). The results evidenced that this alternative medium is suitable for producing higher amounts of Azospirillum brasilense C16 protein and dry biomass in 24 h, allowing for a biological inoculant design with higher cell amounts that ensure plant colonization, as Bashan et al. (2011) reported.
CONCLUSION
When high yields of biomass are required, the substitution of either carbon or nitrogen sources may be not enough to optimize production. Our optimization strategy proved to be an interesting tool to set levels and combinations of sources that maximize biomass production. This alternative culture medium, designed de novo with a sequential statistical strategy, selected and optimized carbon, nitrogen, and minor nutrient sources, allowing the increasing of both protein and dry weight biomass amounts when compared to DYGS. Our alternative medium proved to be efficient for bioinoculant production based on A. brasilense C16. To our knowledge, this is the first study to report the use of sequential statistical designs to obtain a suitable culture medium for mass multiplication of this PGPB.
BIBLIOGRAPHIC REFERENCES
Arzanesh MH, Alikhani HA, Khavazi K, Rahimian HA, Miransari M. 2011. Wheat (Triticum aestivum L.) growth enhancement by Azospirillum sp. under drought stress. World J Microbiol Biotechnol 27:197-205. [ Links ]
Baldani JI, Krieg NR, Baldani VLD, Hartmann A, Döbereiner J. 2005. Genus II. Azospirillum. In: Brenner DJ, Krieg NR, Garrity GM, editors. Bergey's manual of systematic bacteriology. Vol II: The Proteobacteria. Part C: The Alpha, Beta, Delta, and Epsilon proteobacteria. 2nd ed. New York: Springer. pp. 7-26. [ Links ]
Bashan Y, de-Bashan LE. 2010. How the plant growth-promoting bacterium Azospirillum promotes plant growth-a critical assessment. Adv Agron 108:77-136. [ Links ]
Bashan Y, Trejo A, de-Bashan LE. 2011. Development of two culture media for mass cultivation of Azospirillum spp. and for production of inoculants to enhance plant growth. Biol Fertil Soil 47:963-969. [ Links ]
Box G, Behnken D. 1960. Some new three level designs for the study of quantitative variables. Technometrics 2:455-475. [ Links ]
Bradford M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248-254. [ Links ]
Cárdenas DM, Garrido MF, Bonilla R, Baldani VL. 2010. Isolation and identification of Azospirillum sp. in Guinea grass (Panicum maximum Jacq.) of the Valle del Cesar. Pasto Forraje 33:285-300. [ Links ]
Díaz-Zorita M, Fernández-Canigia MV. 2009. Field performance of a liquid formulation of Azospirillum brasilense on dryland wheat productivity. Eur J Soil Biol 45:3-11. [ Links ]
Dobbelaere S, Okon Y. 2007. The plant growth promoting effect and plant responses. In: Elmerich, C, Newton, WE, editors. Associative and endophytic nitrogen-fixing bacteria and Cyanobacterial associations. Dordrecht, The Netherlands Kluwer Academic Publishers. pp. 1-26. [ Links ]
Hartmann A, Bashan Y. 2009. Ecology and application of Azospirillum and other plant growth-promoting bacteria (PGPB)-special issue. Eur J Soil Biol 45:1-2. [ Links ]
Liu C, Liu Y, Liao W, Wen Z, Chen S. 2003. Application of statistically-based experimental designs for the optimization of nisin production from whey. Biotechnol Lett 25:877-882. [ Links ]
Mendes AS, de Carvalho JE, Duarte M, Durán N, Bruns RE. 2001. Factorial design and response surface optimization of crude violacein for Chromobacterium violaceum production. Biotechnol Lett 23:1963-1969. [ Links ]
Peña C, Campos N, Galindo E. 1997. Changes in alginate molecular mass distributions, broth viscosity and morphology of Azotobacter vinelandii cultured in shake flasks. Appl Microbiol Biotechnol 48:510-515. [ Links ]
Plackett R, Burman J. 1946. The design of optimum multifactorial experiments. Biometrika 33:305-325. [ Links ]
Ren J, Lin WT, Shen YJ, Wang JF, Luo XC, Xie MQ. 2008. Optimization of fermentation media for nitrite oxidizing bacteria using sequential statistical design. Bioresour Technol 99:7923-7927. [ Links ]
Spaepen S, Vanderleyden J, Remans R. 2007. Indole-3-acetic acid in microbial and microorganism-plant signalling. FEMS Microbiol Rev 31:425-448. [ Links ]
Wang FQ, Gao CJ, Yang CY, Xu P. 2007. Optimization of an ethanol production medium in very high gravity fermentation. Biotechnol Lett 29:233-236. [ Links ]













