INTRODUCTION
Colombia is one of the main producers of granadilla or passion fruit Passiflora ligularis Juss worldwide, with a production of 51111t during 2020, represented by 4184ha cultivated throughout the national territory, with an average yield of 12.21t ha-1 year-1. The department of Nariño is the fourth largest producer nationwide, in 2020, 970ha of passion fruit were cultivated, with an annual production of 8157t and an average yield of 8.4t ha-1 (MADR, 2022).
Passion fruit production is seriously hampered by vascular wilt caused by a suit of Fusarium oxysporum species. Vascular wilt is the most important pathological problem in passion fruit. In the 90's, there was devastation that occurred in Urrao, Antioquia (Tamayo, 1999). In Brazil, 100% losses were reported due to this disease (Silva et al., 2013).
Geiser et al. (2013) established a single name for the Fusarium species, and through phylogenetic trees grouped them into species complexes. Within the complex of species of Fusarium oxysporum, are the pathogens of plants, animals, and humans; as well as a wide range of non-pathogenic individuals that are used in biological control (Gordon, 2017).
Fusarium is able to survive for a long time in soil and plant residues the fungus generally penetrate naturally or also require wounds to enter the plant and colonize the tissues (García et al., 2007). These wounds can be bark natural cracks, wounds caused by insects, nematodes, or by humans during the crop management. When this pathogen attacks in the early stages, the seedlings display yellowing poor growth. In later stages it eventually causes plant death (Tamayo, 2001; Melgarejo & Rodríguez-Castillo, 2015).
In Colombia, some studies had been carried out to identify the causal agent of vascular wilt in Passiflora ligularis. Molecular characterization of Fusarium spp. associated with Passiflora edulis in Valle del Cauca, showed that the species that caused this disease were F. incarnatum, F. proliferatum, and F. solani, where F. incarnatum is the strain with the highest occurrence in the evaluated samples. F. oxysporum and F. solani have been identified as the causal agents in Passiflora edulis Sims in the Sumapaz region (Ortiz, 2012; Henao et al., 2017).
Molecular approaches based on the comparison of DNA sequences are currently the most used for the study of population genetics, taxonomy, phylogeny, and systematics of Fusarium spp. Multi-loci sequencing, such as translation elongation factor 1α (EF-1α, TEF) and ITS regions of ribosomal DNA has been used for molecular systematics (Balajee et al., 2009; Salazar, 2016).
The establishment of vascular wilt management and control strategies required adequate knowledge at the taxonomic and phylogenetic level of the organisms that cause the disease (Alastruey-Izquierdo et al., 2008; Landlinger et al., 2009; Sampietro et al., 2010).
Hence, the present study focuses on the identification of fungal isolates that might be associated with vascular wilt in passion fruit (P. ligularis) crops of South of Colombia.
MATERIALS AND METHODS
Location and sampling. The study was carried out in the laboratory of the Grupo de investigación en Sanidad Vegetal (GRISAV) of Universidad de Nariño, located at 2488 masl, 1°14´00.89” N and 77°17´38.66 ”W, and a temperature of 14°C.
Isolates were obtained from symptomatic P. ligularis displaying symptoms such as chlorosis or yellow leaves, stem necrosis, and wrinkling of the fruits (García et al., 2007). The sampling was randomly carried from six farms in the municipalities of Ancuya, San Pablo, and Guaitarilla in the department of Nariño (Colombia). Roots and stems Symptomatic and asymptomatic plants were sampled and packed in hermetic plastic bags.
Fungal isolation. The isolation process was carried out by direct platting on the media of the affected tissue, following the procedure described by Leslie & Summerell (2006). In short, under a laminar flow chamber, and with a sterile scalpel, diseased and healthy tissue sections were made. Samples were surface sterilized by immersion into sodium hypochlorite 2.5% (NaOCl) for one minute, then in 70% ethanol for one minute, and finally an intense rinse with sterile distilled water was carried out. In order to obtain pure isolates generated from a single spore; a suspension of low concentration of conidia in sterile water was obtained, by means of dilutions, from the initial culture.
Morphological identification. The isolates were characterized under the microscope considering traits such as colony growth per day (gpd), and color. For colony growth diameter in eight days and were grouped taking into account the growth per day (gpd) categories: slow (<10 mm gpd), medium (> 10-12 mm of gpd) and rapid (> 12 mm gpd) (Dubey et al., 2010). The mycelium color was evaluated by comparing the pigmentation presented on the front and back of the Petri dishes with the color catalog (The online Auction Color Chart, 2004). The microscopic parameters analyzed were macroconidia shape (basal cell and apical cell), microconidia, presence or absence of chlamydospores, and phialides (Leslie & Summerell, 2006).
Molecular characterization. DNA extraction was performed by the method of Caetano et al. (2015) with some modifications. Thus, in each tube with mycelium, 500 µl of buffer solution was added (250mL Tris-HCl100 mM pH 7.2; 250mL EDTA100 mM pH 8 and 250mL sodium dodecyl sulfate (SDS) at 10%, pH 7.2). Subsequently, a maceration was carried out using sterile micropipette tips, in order to break the cell walls. Tubes were then incubated at 65°C for one hour in a water bath (Memmert WNB 7). After the incubation, 500µl phenol/chloroform/isoamyl alcohol was added to each tube, in a ratio of 25:24:1, tubes were shaken by inversion for five minutes and centrifuged at 13000 rpm for 12 minutes, in a centrifuge (HERMLE Z 216M). The supernatants were transferred to microtubes, to which 500µl of chloroform/isoamyl alcohol (24:1) were added per tube; subsequently, they were shaken by inversion for five minutes and centrifuged at 13000 rpm for 12 minutes. The supernatants were transferred to microtubes, then 50µl of sodium acetate and 800µl of cold absolute ethanol were added to precipitate the DNA. The tubes were left at -20°C overnight. After they were centrifuged at 13000rpm for 12 minutes, the supernatant was removed and the pellet contained in the lower phase of the tube was allowed to dry at room temperature for two hours in the laminar flow chamber. Finally, the DNA was diluted in 30 µl of milli q water, to then determine its integrity by visual appreciation in agarose gel (0.6%). Subsequently, it was stored at -20°C until its use to amplify the ITS region and TEF1α.
ITS region of rDNA was amplified with the universal primers described by White et al. (1990); Salazar (2016); and Salazar et al., (2020)ITS1 (5'-TCCGTAGGTGAACCTGCGG-'3) and ITS4. (5'TCCTCCGCTTATTGATATGC-3 '), synthesized by Macrogen, Korea. The mixture for the amplification was carried out in a volume of 30µl, 15µL of Green Master mix (Biolabs ®) was added, 1µL of each primer (10µM), 3µL of DNA were added and it was completed with nucleases-free water. Amplification was performed in a thermal cycler (Labnet MultiGene ™ OptiMax, 96 well programmable). The temperature cycles were 95°C for one minute, to denature the samples, 35 denaturation cycles at 95°C for 30 seconds, 55°C for one minute for alignment, an extension of 72°C for 45 seconds, and finally, elongation at 72°C for five minutes and termination at 4°C for five minutes.
The TEF Ef1α and Ef2α gene region was amplified with the 5'ATGGGTAAGGAGGACAAGAC-3 and 5'-GGAGGTACCAGTGCATCATGTT-3 primers, synthesized by Macrogen, Korea (O'Donnell et al., 1998; Salazar, 2016).
The PCR reaction was performed in a final volume of 30µl which contained 15µl of Green Master mix (Biolabs ®) then was added 1µl of each primer, 3µl 100ng of DNA, and it was completed with the nuclease free, water contained in the kit. The PCR program comprised an initial denaturation at 95°C for 2 minutes, followed by 35 cycles of denaturation at 95ºC for 30 seconds, hybridization at 56°C for 30 seconds, an extension of 72°C for one minute, and elongation finish at 72°C for 10 minutes and finish at 4°C for 5 minutes. Amplifications were carried out in a thermal cycler (Labnet, MultiGene ™ OptiMax, 96 well programmable).
A 100bp molecular weight marker (Biolabs ®) was included on a 1.2% agarose gel, developing with HydraGreenTM (HydraGene), and then this gel was run at 70 Volts for 40 minutes in a horizontal electrophoresis chamber. The gels obtained were analyzed in a transilluminator (Fotodyne 3-3500 Photo / PrepI UV) to determine the amplification of the expected band in the ITS and TEF 1α regions.
PCR products were isolated and sent for sequencing using the Sanger method in the Macrogen, Korea laboratory and the Basic Local Alignment Search Tool (National Library of Medicine, 2021) software from NCBI (National Center for Biotechnology Information) was used for the species comparison.
Phylogenetic analysis. The sequencing results were edited with the BioEdit software (version 7.0.5.3). After editing the sequences, a multiple alignments was performed by Clustal W, using the MEGA X computer program (version 10.0). With the same program and from the multiple alignments, the phylogenetic tree was built, using the similarity coefficient of the nearest neighbor (Lin et al., 2014) with a Bootstrap of 1000 replicas.
Trichoderma sp. sequences (Accession MF185948.1) for TEF 1 α and (Accession N ° MK108405.1) for ITS, deposited at the database of GenBank, were selected to have a reference species in the phylogenetic tree construction.
RESULTS AND DISCUSSION
Thirty-five isolates were successfully gattered from the samples, 23 from the municipality of San Pablo, six from the municipality of Ancuya, and six from the municipality of Guaitarilla.
Morphological characterization. At the macroscopic level, in the 35 isolates, diversity was observed in terms of colony color, displaying a cottony appearance and a whitish-pinkish to purple-red color in the first stages of development (Figure 1). Subsequently, as the mycelium developed, it turned a slightly purple color in the center and others tuned into a more intense red color. On the reverse side of the Petri dish, a dark purple color was observed towards the center in most isolates and a lighter tone towards the outer edge, these results agree with those recorded by Retana et al. (2017) such of Fusarium species or oxysporum characteristics. In addition, in one isolate a cottony texture colony was observed, with aerial mycelium in the center and a brown to cinnamon color, characteristics associated with Ilyonectria sp. as mentioned by Pathrose (2010).
The colony presented rapid growth (>12 mm gpd), in 29 isolates; this agrees with studies carried out by Rodríguez (2013) as a characteristic of Fusarium oxysporum, five isolates with a medium growth (10-12mm of growth per day), corresponding to SP19, SP21, SP22, AN24 and AN26 and isolation (GU32), with low growth (less than 10mm growth per day).
Under the light microscope in 34 isolates the typical form of Fusarium oxysporum, although, with some differences between them, since some were slightly shorter and others less curved. On average, macroconidia had three to five septa, with a rounded apical cell and a slightly curved basal cell (Figure 1A). In the case of microconidia, the oval shape was more commonly observed, in most of them without septa (Figure 1B), short monophysites were also formed (Figure 1C). In addition, smooth-walled chlamydospores were observed, in a simple intercalary or terminal manner, in pairs or chains (Figure 1D), and the formation of microconidia growing in false heads (Figure 1E). These results were compared to those obtained by Nelson et al. (1983).
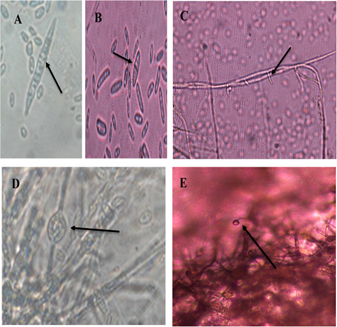
Figure 1 Morphological structures of Fusarium oxysporum. A. Septate macroconidia, with elongated and slightly curved apical and basal cells. B. Septate oval microconidia. C. Short monofialides. D. Smooth-walled chlamydiospores. E. Microconidia growing on false heads.
Likewise, in one isolate, straight, hyaline macroconidia were observed, with a basal prominence and with no more than three septa (Figure 2A) and microconidia with an ellipsoid to an oval shape, which had from zero to one septum (Figure 2B), growing in false heads (Figure 2C). Abundant chlamydospores were also observed, growing in chains, of brown color (Figure 2D), with appearances of Ilyonectria sp. according to Chaverri et al. (2011); Iturralde, (2017).
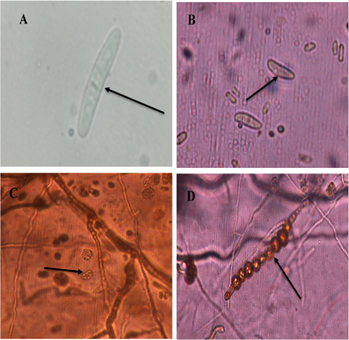
Figure 2 Morphological structures of Ilyonectria sp. A. Septate macroconidium. B. Oval microconidia. C. Microconidia growing on false heads. D. Globous, chain-like Chlamydiospores with thick walls.
These results are similar to those from Patiño et al. (2015), and Osorio et al. (2020), who mention that the diagnosis of the disease known as secadera is related to the species of F. oxysporun fs passiflorae and F. solani f sp. passiflorae being the species F. solani the one with the highest pathogenicity in P. ligularis plants In Colombia, through the sequencing of the intergenic spacer (IGS) rDNA, obtaining that the species causing this disease were Fusarium oxysporum and Fusarium solani (Patiño et al., 2015)
In addition, Londoño (2012) carried out the molecular characterization of Fusarium isolates associated with the passion fruit (P. edulis) vascular wilt in Valle del Cauca, obtaining that Fusarium oxysporum was among the causes of this disease; also showing that within the F. oxysporum cluster, other subgroups were formed, which demonstrated that there was intraspecific variability. It is considered that the source of variability for this species that does not present sexual reproduction, would be associated with mutations This phenomenon facilitates gene transfer via hyphal anastomosis and the presence of transposon elements; which allows asexual reproduction to establish clonal lineages within the population (Jacobson & Gordon, 1991; Salazar, 2016). Transposon elements act as diversity generators because their activity produces spontaneous mutations and consequently variability in the genome (McDonald et al., 1992; Daboussi & Langin, 1994).
Sequencing of the ITS and TEF-1α region. For the ITS region, each of the 35 isolates was identified, of which 34 were associated with Fusarium oxysporum and one with Ilyonectria robusta. Interstingly, it was observed in 22 isolates with 100% identity, 11 isolates presented a percentage greater than 95% and two isolates presented values of 77 and 85% identity (Table 1).
Basic Local Alignment Search Tool (BLAST) analysis for TEF-1α region in the 35 isolates resulted in 34 isolates belonging to Fusarium oxysporum. Of these, 14 isolates presented a special form of Fusarium oxysporum f. sp. passiflorae and one isolate belonged to Ilyonectria robusta. 26 isolates presented a high level of identity with a percentage greater than 98%, six isolates presented a percentage greater than 95% and three isolates presented identity values of 85, 90, and 91%.
Table 1 Identification at the species level by analysis of percentage similarity and accession code (according to GenBank) of the sequences obtained in the ITS region (Internal transcribed spacer) and TEF-1α (Translation elongation factor).
| Code | ITS | Accession | % | TEF | Accession | % |
|---|---|---|---|---|---|---|
| SP1 | Fusarium oxysporum | MK246922.1 | 100 | Fusarium oxysporum | MH582354.1 | 99 |
| SP2 | Fusarium oxysporum | MK271759.1 | 100 | Fusarium oxysporum f. sp. passiflorae | JF332039.1 | 98 |
| SP3 | Fusarium oxysporum | MK246922.1 | 100 | Fusarium oxysporum f. sp. passiflorae | JF332039.1 | 96 |
| SP4 | Fusarium oxysporum | MG251531.1 | 98 | Fusarium oxysporum f. sp. passiflorae | KX434919.1 | 99 |
| SP5 | Fusarium oxysporum | MK212364.1 | 100 | Fusarium oxysporum | MH582354.1 | 97 |
| SP6 | Fusarium oxysporum | MH752510.1 | 100 | Fusarium oxysporum | KU361438.1 | 99 |
| SP7 | Fusarium oxysporum | MK246922.1 | 100 | Fusarium oxysporum | MF327628.1 | 90 |
| SP8 | Fusarium oxysporum | MH221085.1 | 100 | Fusarium oxysporum | MH401561.1 | 99 |
| SP9 | Fusarium oxysporum | MK246922.1 | 100 | Fusarium oxysporum | KY657530.1 | 99 |
| SP10 | Fusarium oxysporum | KY910856.1 | 100 | Fusarium oxysporum | EU091046.1 | 100 |
| SP11 | Fusarium oxysporum | MH115588.1 | 100 | Fusarium oxysporum f. sp. passiflorae | JF332039.1 | 98 |
| SP12 | Fusarium oxysporum | MK239978.1 | 100 | Fusarium oxysporum | MH172303.1 | 100 |
| SP13 | Fusarium oxysporum | MG975622.1 | 95 | Fusarium oxysporum f. sp. passiflorae | FJ985362.1 | 99 |
| SP14 | Fusarium oxysporum | LS479732.1 | 95 | Fusarium oxysporum | MH582353.1 | 99 |
| SP15 | Fusarium oxysporum | GQ121320.1 | 77 | Fusarium oxysporum f. sp. passiflorae | FJ985362.1 | 99 |
| SP16 | Fusarium oxysporum | KU982608.1 | 99 | Fusarium oxysporum f. sp. passiflorae | JF332039.1 | 100 |
| SP17 | Fusarium oxysporum | MH913119.1 | 100 | Fusarium oxysporum f. sp. passiflorae | KX434919.1 | 99 |
| SP18 | Fusarium oxysporum | MH115588.1 | 100 | Fusarium oxysporum f. sp. passiflorae | KX434919.1 | 99 |
| SP19 | Fusarium oxysporum | MK246922.1 | 99 | Fusarium oxysporum | MH401561.1 | 99 |
| SP20 | Fusarium oxysporum | MK212364.1 | 100 | Fusarium oxysporum | HQ731058.1 | 91 |
| SP21 | Fusarium oxysporum | MG798785.1 | 100 | Fusarium oxysporum | MH582354.1 | 97 |
| SP22 | Fusarium oxysporum | KX823406.1 | 100 | Fusarium oxysporum | EU091046.1 | 85 |
| SP23 | Fusarium oxysporum | MK156696.1 | 100 | Fusarium oxysporum | KX215038.1 | 96 |
| AN24 | Fusarium oxysporum | MK204578.1 | 100 | Fusarium oxysporum | MH119941.1 | 96 |
| AN25 | Fusarium oxysporum | MK185381.1 | 100 | Fusarium oxysporum f. sp. passiflorae | KX434919.1 | 99 |
| AN26 | Fusarium oxysporum | MF280112.1 | 100 | Fusarium oxysporum f. sp. passiflorae | JF332039.1 | 99 |
| AN27 | Fusarium oxysporum | MG407704.1 | 96 | Fusarium oxysporum | HQ731058.1 | 99 |
| AN28 | Fusarium oxysporum | MH752510.1 | 100 | Fusarium oxysporum | JF794777.1 | 96 |
| AN29 | Fusarium oxysporum | JN859839.1 | 99 | Fusarium oxysporum f. sp. passiflorae | KX434919.1 | 99 |
| GU30 | Fusarium oxysporum | MG975622.1 | 99 | Fusarium oxysporum f. sp. passiflorae | KX434919.1 | 99 |
| GU31 | Fusarium oxysporum | MG975622.1 | 98 | Fusarium oxysporum f. sp. passiflorae | JF332039.1 | 98 |
| GU32 | Ilyonectria robusta | KY555034.1 | 99 | Ilyonectria robusta | JF735717.1 | 99 |
| GU33 | Fusarium oxysporum | MG272267.1 | 85 | Fusarium oxysporum | LS423229.1 | 99 |
| GU34 | Fusarium oxysporum | KT223907.1 | 99 | Fusarium oxysporum | MF327621.1 | 98 |
| GU35 | Fusarium oxysporum | MH752510.1 | 100 | Fusarium oxysporum | KY657530.1 | 98 |
Phylogenetic analysis of the ITS Region. From the phylogenetic tree generated, two clearly differentiated clusters can be observed, those clades correspond to the species Fusarium oxysporum and Ilyonectria robusta (GU32) (Figure 3). In addition, within the species, there are four clades. The isolates (clade I, monophyletic) twenty-eight reflect that there is no genetic variability. The minimum distances between the species are found, while with the individuals that are at the end of the tree there is high diversity and divergence between the sequences of isolates evaluated.
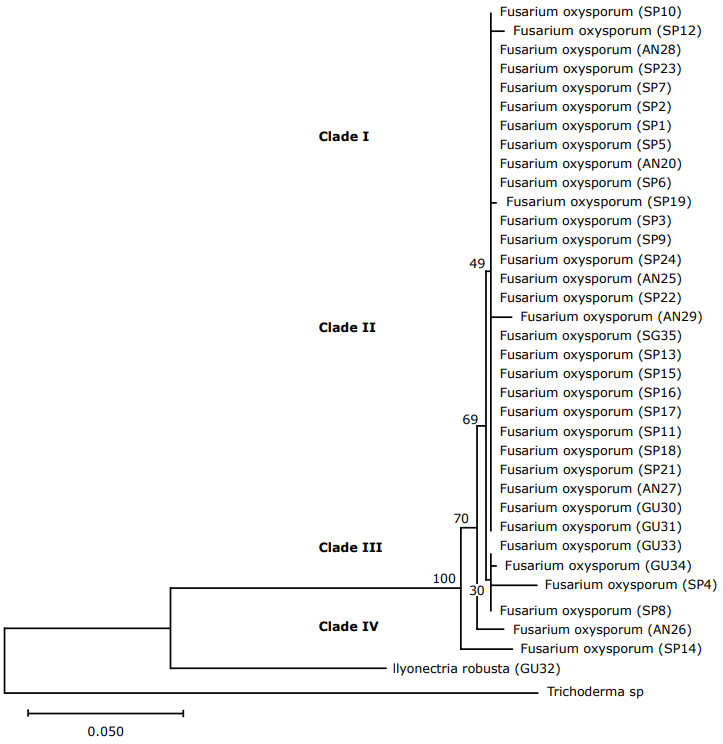
Figure 3 Phylogenetic tree of Fusarium sequences with the first ITS 1-ITS 4 (Internal transcribed spacer) using the Neighbor joining NJ similarity coefficient. The tree was built with a 1000 replica bootstrap. (SP: San Pablo; AN: Ancuya; GU: Guaitarilla).
Also, it can be seen that in the first part of the tree, there are the minimum distances between the evaluated isolates, showing a wide similarity within the group, which indicated that most of these came from a common ancestor due to the homology of their sequences (Geiser et al., 2013). While in the individuals that are at the end of the tree there is high diversity and divergence. In clade I and II it is observed that most isolates are of monophyletic origin; however, the isolates of clade III could be related to non-pathogenic isolates and show these differences in the sequences analyzed.
The ITS region was insufficient for the resolution of groups because it was also reported for closely related species within the Fusarium genus (Turner et al., 1998). The species Ilyonectria robusta is found in this area, a teleomorph of Cylindrocarpon destructans that, according to the classification of Aoki et al. (2012) is not found within the Fusarium group, agreeing with its position in the phylogenetic tree.
TEF 1α Region Analysis. In the TEF1α primer, 34 isolates were determined and classified as Fusarium oxysporum (Figure 4) (clade I and II) coming from Ancuya, San Pablo, and Guaitarilla municipalities, and one Ilyonectria robusta (GU32), coming from Guaitarilla municipality.

Figure 4 Phylogenetic tree of Fusarium sequences with the first TEF-1α (Translation Elongation Factor) using the Neighbor-joining NJ similarity coefficient. The tree was built with a 1000 replica bootstrap. (SP: San Pablo; AN: Ancuya; GU: Guaitarilla).
Most studies use the tef-1α gene to establish phylogenetic relationships within and between intraspecific groups (Nalim et al., 2009). In this case, it is observed that clade I isolate could be related to non-pathogenic Fusarium oxysporum, which would be verified in the next study through pathogenicity tests.
In the phylogenetic tree of the TEF 1α region, as in the ITS region, two clear clusters associated with Fusarium oxysporum and Ilyonectria robusta species were obtained. However, in this particular case it can be noted that for the Fusarium oxysporum group, isolates with the special form Passiflorae were placed.
CONCLUSIONS
The species Fusarium oxysporum was the fungus isolated in the three producing municipalities of Passiflora ligularis in the department of Nariño and is related to the causal agent of vascular wilt in its special form Fusarium oxysporum f.sp. passiflorae. The morphological characteristics of the isolates were correlated with those reported for this species.
The Illonectria robusta species is a teleomorphic phase of Cylindrocarpon destructans, which has not been reported so far as a pathogen in any species of passiflorae. Therefore, it is necessary to carry out pathogenicity tests to determine how it can affect sweet passion fruit plants.
The Phylogenetic analysis allowed determining that there was little diversity among the evaluated isolates, not finding any variation in relation to the geographical origin of the samples. This may have been due to the use of a single ecotype among producers, called passion fruit.















