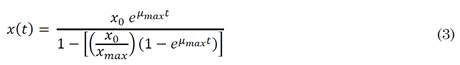1. INTRODUCTION
Lactic Acid Bacteria (LAB) are present in a wide range of artisanal products, such as fermented foods and beverages [1]. They are continuously used in the dairy industry because of their metabolic characteristics, such as acidifying activity, resistance to bacteriophages, proteolytic activity, synthesis of bacteriocins, and production of exopolysaccharides, properties that allow them to be recognized with GRAS (Generally Recognized As Safe) status, contribute to the development of flavor and texture, and also provide nutritional attributes in this type of product. Nevertheless, some LAB strains must be evaluated for their safety properties such as antimicrobial resistance and virulence genes, before industrial use [2], [3]. Milk and artisanal dairy products, such as cheese, constitute an important source of indigenous LAB [3], [4] which can be used at an industrial level to explore and apply natural biodiversity to provide a suitable means to promote the quality and safety of new products or the improvement of existing processes [5].
The use of autochthonous LAB for industrial purposes, such as the formulation of starter cultures for use in pasteurized milk, implies its technological characterization, which includes the production of lactic acid and other metabolites such as acetic acid, amino acids, fatty acids, exopolysaccharides, vitamins, acetoin, acetaldehyde, mannitol, bacteriocins, and antioxidants, and seeks to preserve the sensory qualities of the original product [6], which could offer nutritional and sensorial characteristics desirable in dairy products such as cheese. This requires a careful selection of media and operating conditions to obtain optimal and reproducible results in terms of final cell number, activity, and stability during storage[7].
The kinetics describe the phases of individual growth, which include, the adaptation phase, exponential phase, stationary phase, and decay phase, which are explained by non-linear kinetic models [8]. These models constitute a fundamental tool to determine the quantitative importance of bacteria, monitor and predict fermentative processes, and improve their analysis for biotechnological use. Likewise, they provide advanced information on the behavior of microorganisms using precise, repeatable, detailed experiments and mathematical models, which contribute to the design and control of biotechnological processes [9].
The kinetics of LAB microbial growth have been studied using mathematical models such as the logistic and Gompertz models, which do not consider the rate of substrate consumption because they assume that the concentration of this remains high enough to reach the maximum number of microorganisms during the culture time [10]. The logistic model relates the growth rate to two kinetic parameters, µmax (maximum specific growth rate), and biomass concentration; in the Gompertz model, the growth rate is proportional to the cell biomass, which decays exponentially with time due to bacterial inactivation [9].
Autochthonous LAB strains have an advantage over allochthonous strains, being more niche specific and allowing the production of competitive high-quality products. There is a constant search for new commercially available autochthonous for the formulation of starter cultures where sensory, nutritional, and technological attributes could be more consistently assured [11]. In Colombia, few studies have been focused on the identification of LAB in autochthonous products, such as lactic ferments [12] and traditional cheeses such as Double Cream, where some of their technological and probiotic properties are partially described[13], without the evaluation of the growth conditions, production of lactic acid, and survival in the substrate, attributes necessary for industrial use. Therefore, the present work was proposed to evaluate in UHT milk, the growth kinetics, the pH decrease, and lactic acid production, of autochthonous LAB previously obtained from a traditional Colombian cheese "Double Cream", to identify its potential in the formation of a starter culture that contributes to improving the traditional production techniques of this cheese using pasteurized milk, without losing the qualities of the original product.
2. METHODS
2.1 Bacterial strains and inoculum preparation
For this study, native strains of LAB isolated in a previous work [14] from Double Cream cheese and identified by partial sequencing of the 16S rRNA gene as Pediococcus pentosaceus (19), Leuconostoc citreum (20), Pediococcus acidilactici (21), Leuconostoc mesenteroides subsp. mesenteroides (22, 29), Enterococcus faecium (24, 25), Enterococcus faecalis (27), Weissella viridescens (28), Lactococcus lactis (30), Lacticaseibacillus casei (31) and Limosilactobacillus fermentum (32) The strains were used under permit 1467 dated December 03, 2014, granted by the ANLA (National Authority for Environmental Licensing).
Strains preserved in 10 % v/v glycerol at -70 °C were activated in MRS broth (Merck) and incubated at 37 °C for 48 h under anaerobic conditions. After this time and independently for each strain, the cell concentrations were adjusted to 0.5 on the McFarland standard, using peptone water, and the inocula were prepared at 10 % v/v in ultra-high temperature (UHT) treated milk, for a final volume of 80 mL. 100 mL Schott flasks were used, and the cultures were left shaking at 120 rpm for 12 h at 37 °C.
2.2 Growth kinetics
80 mL of the inoculum was transferred to 1000 mL glass flasks containing 720 mL of UHT milk, the cultures were shaken at 120 rpm at 35 - 37 °C, until the milk coagulated. Every 3 hours, 8 mL samples were taken to determine cell growth, pH, and acidity.
2.2.1 Quantification of cell growth
From each of the samples, serial dilutions were prepared, and the pour plate method was performed in MRS agar for counting the colony-forming units (CFU/mL). Cultures were incubated at 35-37 °C for 48 h under anaerobic conditions. The results were expressed in logarithmic units over time (Log10 CFU/mL).
2.2.2 Measurement of pH and titratable acidity
The pH was measured using a potentiometer (330 Super Scientific), and the total titratable acidity (TTA) was evaluated up to pH 8.2 using the AOAC (2000) 939.05 [15] method with 0.1 N NaOH and expressed in percent acidity (g/100 mL).
2.3 Determination of kinetic parameters
The exponential model was used to fit the experimental data obtained during the growth kinetics. Equations (1) and (2) were used to compare specific growth rates (µ x ) and doubling time (T d ), respectively. Logarithmic growth phase data were used and fitted by linear regressions using Polymath 5.1.
Where x corresponds to CFU/mL, t is the time in hours and x 0 CFU/mL at time 0, T d is the doubling time, Ln 2 corresponds to the natural logarithm of 2 and µ x is the specific growth rate. The logistic model was used to estimate the kinetic parameter, maximum growth rate (µ max ) for each of the evaluated strains, according to equation (3).
Where x(t) corresponds to the CFU/mL as a function of time, x 0 to the CFU/mL at time 0, and µ max the maximum specific growth rate [16]. The model was validated by using the average of three fermentation batches. The coefficient of determination (R2) was used to validate both the exponential and logistic models.
2.4 Statistical analysis
All experiments were performed independently in triplicate and data were represented as the mean ± standard deviation. For the analysis of the experimental data (pH and titratable acidity) the Kruskal Wallis test for median differences was used. All statistical analyses were performed with a confidence level of 95 %.
3. RESULTS AND DISCUSSION
3.1 Growth kinetics
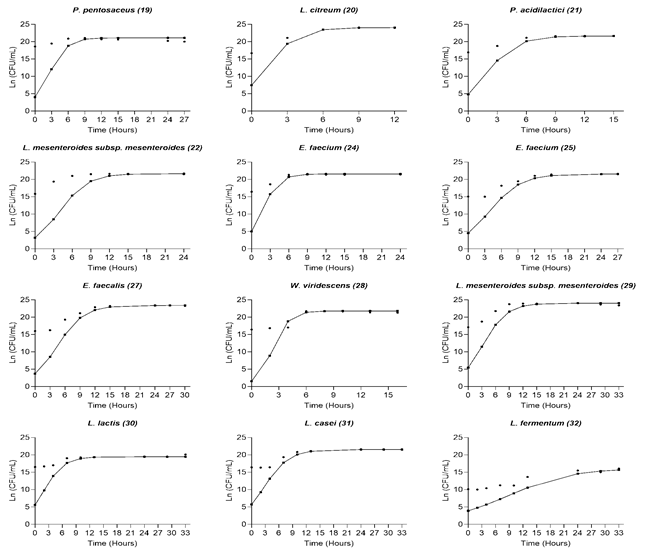
The shake flasks used for the kinetics constituted a primary and rapid system for the detection and optimization of microbial growth before leading to a bioreactor scale-up; gentle shaking in this type of flask did not slow down the growth of the microorganisms studied[17].Figure 1 shows the growth curve and the percentage of acidity of each of the strains studied. The latency phase lasted between 0 and 5 hours, and the exponential growth phase lasted between 8 and 15 hours; after that, the strains entered the stationary phase. The bacterial concentration increased during the kinetics between two and three logarithmic units, starting at 7 Log10 CFU/mL. For most of the strains, during the exponential phase, the highest production of organic acid occurred.
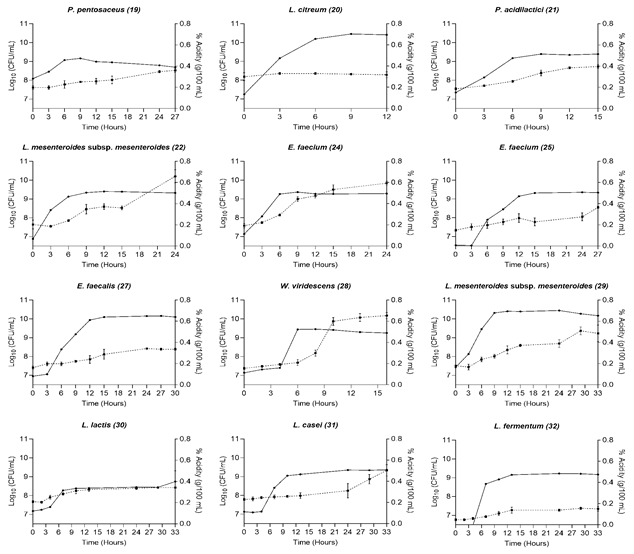
Source: Created by the authors.
Figure. 1 Bacterial growth and titratable acidity (-●) Log10 (CFU/mL), (--▪) % Acidity (g/100mL)
A short latency phase accounts for the good adaptation that microorganisms have to the substrate used [18] since it shows a rapid activation of signaling pathways and transcriptional changes that positively regulate the metabolism of bacteria, a necessary process for their differentiation and multiplication in the exponential phase [19]. In this study, the genera Lactococcus, Pediococcus, and Leuconostoc presented the shortest 0-2 h latency phases in UHT milk used as substrate. For L. lactis, rapid growth in milk has been reported because its aerobic metabolism leads to a more energetic balance with one more ATP molecule produced than under anaerobic conditions [20]. However, this behavior has also occurred when using selective media as a substrate [21]. In milk, the amino acid concentration does not allow for the growth of Lc. lactis above 8 log (CFU)/mL. To continue its development, the bacterium needs to use the caseins and activate its proteolytic metabolism, which could become a limiting factor for its growth in this substrate [22]. The genus Pediococcus has shown short adaptation phases in fermented products such as soy milk and selective culture media [23], [24]. On the contrary, for the genus Leuconostoc, adaptation phases that exceed 6 h have been reported using the same substrate as in the present study, at a temperature of 30 °C [25]. However, it has also been described that temperature is a decisive factor in the growth of this microorganism, which exhibits its best growth rate between 25-30 °C and an increase in it can decrease its adaptation phase[26], [27].
Lactobacilli have been used as starter cultures in dairy fermentative processes due to their optimal growth in milk and ability to produce metabolites that contribute to product quality and confer health benefits [28]. In our study, the strains L. casei (31) and L. fermentum (32) presented adaptation phases between 0-4 h, being higher for L. casei, which coincides with the behavior reported by [8] for these species and even had a shorter duration than that reported in other studies [29]. The time required for lactobacilli to reach adequate adaptation of their enzymatic machinery to carry out lactose hydrolysis explains the difference in the duration of the adaptation phase with Lactococcus, Pediococcus, and Leuconostoc [29].
Enterococcus is one of the most abundant genera within the natural microbiota of milk; may play an important role in the development of aromas of fermented products such as cheese [30]. In this study, the strains E. faecium (25) and E. faecalis (27) presented adaptation phases higher than 4 hours, according to what was indicated in other reports [31]. E. faecium (24) presented a shorter adaptation phase, indicating that the growth of these microorganisms is variable between strains and related to their adaptation to the substrate [30]. Finally, Weissella viridescens (28) presented an adaptation phase of close to 4 h and a short exponential phase of 4-8 h, findings that agree with other studies [32] and corroborate its good behavior in milk.
3.2 pH and titratable acidity (TTA)
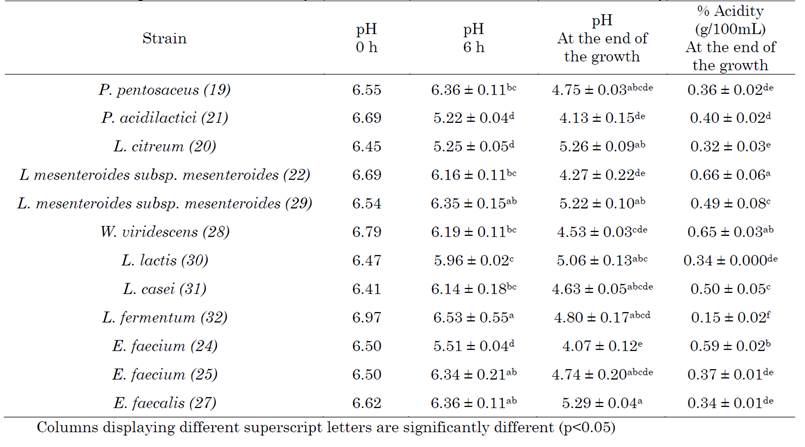
The decrease in pH values due to the production of lactic acid plays an important role in the production processes of dairy products such as cheese, since it favors the production of components related to organoleptic properties and leads to the growth inhibition of some pathogens [33]. During the kinetics, there was a decrease of two units in the pH values of the medium, starting at 6.5 and reaching values of 4.07 (Table 1). One of the most important properties of the candidate strains for the formulation of starter cultures used in milk is their ability to reduce the pH from values of 6.6 to 5.3 in the first 6 hours of growth [34]. In this study, the strains P. acidilactici (21), E. faecium (24) and L. citreum (20) (Table 1) showed this characteristic. However, when talking about autochthonous strains, it is considered that the decrease in pH to values below 5.1 at the end of the kinetics is sufficient to consider their use in starter cultures [35]. The best acidifiers were P. acidilactici (21), E. faecium (24), and L. mesenteroides (22), behavior that was found in the majority of our strains (Table 1).
P. acidilactici has been reported as a strain that produces acidity from the fermentation of sugars, in addition to presenting coagulant activity, qualities that have suggested it as a representative of starter cultures [36]. On the other hand, it has been described that enterococci show a low or medium capacity to reduce the pH of milk, with some exceptions such as E. faecium, which has been shown to have a good acidification capacity in dairy matrices [37], which coincided with what was observed in this work. Other enterococci strains E. faecalis (27) and E. faecium (25) showed a lower reduction, both at 6 h and at the end of the kinetics. The pH for the Leuconostoc strains (20-22-29) at the end of the growth kinetics stabilized in a range between 4.27 and 5.26, data that corresponds to other reports for this bacterial genus [35], in which it is described that Leuconostoc strains, derived from dairy products, adapt very well to acidic environments. However, L. citreum, which in the first 6 hours showed a significant drop in pH, failed to pass the threshold of 5.1 at the end of the kinetics, which could be explained by the presence of organic acids in the medium that can have an inhibitory effect on the growth of Leuconostoc and cause an early entry into the stationary phase [25], [38].
In our study, the lactobacilli decreased the pH between 6.3 and 6.6 in the first 6 hours of kinetics, reaching values of 4.7 after 24 hours (Table 1); This result coincides with what was reported in another study [39] that evaluated the acidification capacity in UHT milk of several strains of lactobacilli isolated from a traditional cheese, it has even been described that some strains are capable of reducing the pH of the milk to values close to 3.0 after 24 hours [40]. The genus Weissella has been reported to present an accelerated growth rate, reaching pH values below 4.2 in the first 24 hours [41], [42], similar to the data presented in this study, where the decrease in pH at 16 hours was around 4.5.
The determination of titratable acidity (TTA) allows for the quantification of organic acid production, with lactic acid being the main metabolic product of LAB at the end of fermentation. The TTA increased by an average of 0.3 g/100 mL, reaching values above 0.6 g/100 mL, at the end of the growth kinetics (Table 1). The acid production rate gradually increased during kinetics, as a result of the metabolism of carbohydrates present in milk and was higher in the exponential phase than in the adaptation phase (Figure 1), as has been reported in other studies performed on LAB [16]. At the end of the kinetics, there were significant differences in the quantification of TTA between the microorganisms (p<0.05) (Table 1), being L. mesenteroides (22), E. faecium (24) and W. viridescens (28), the strains that presented the highest percentage of acidity with values between 0.59 and 0.66 g/100 mL and that, in turn, were able to reduce the pH below 4.5; a characteristic that has been described in other studies for these species [43]. Strains of Leuconostoc mesenteroides from dairy environments are characterized by their good ability to produce acidity [44]; however, their use in starter cultures is conditioned by their contribution to flavor production, either when used alone or in combination with other strains such as L. lactis, thanks to its hetero-fermentative character and its ability to metabolize the citrate present in milk to generate aroma by the formation of diacetyl [45]. Some Enterococcus species have the ability to produce high amounts of lactic acid, which contributes to the rapid acidification of milk and the production of flavor and texture [46]. As observed in Table 1, E. faecium (24) presented a higher % acidity than E. faecalis (27), a behavior that has already been reported in other studies for these two species [47]. W. viridescens, due to its good acid production and probiotic properties [29], has been reported as a member of starter cultures for fermented products, and despite being a heterofermentative microorganism, it has shown greater production of lactic than acetic acid in these processes [32].
Lactobacillus and Lactococcus are commonly described as lactic acid producers [3]. In our study, L. casei (31) presented a higher percentage of acidity than L. fermentum (32), in agreement with another study carried out on UHT milk [43], which highlighted for these two species their good ability to produce flavor precursor secondary metabolites. On the other hand, L. lactis, a homolactic bacterium with a good rate of lactic acid production, which has been considered a member of starter cultures for many fermented products, mainly cheeses, did not show a good response in this study (Table 1), which may be due to an affectation of its glycolytic rate produced by the tension of the pH of the medium [18].
3.3 Determination of growth parameters
Kinetic models play an important role in monitoring and predicting fermentative processes involved in substrate utilization, growth, and product formation, and are recognized as an informative tool for rapid and cost-effective evaluation of microbial growth[9]. The doubling time is related to the growth specific rate, and both parameters can be used to describe changes in the number of microorganisms over time [48]. The kinetic parameters for the strains in this study are described in Table 2.
Columns displaying different superscript letters are significantly different (p<0.05).
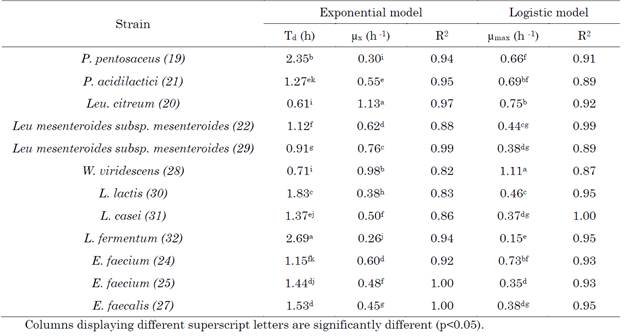
As shown in Table 2, there were significant differences (p<0.05) in the kinetic parameters µx and Td obtained with the exponential model for the studied strains, namely L. citreum (20), L. mesenteroides (29) and W. viridescens (28), which showed the shortest doubling times and the highest specific growth rates (between 0.76 and 1.13 h-1). These data are similar to those presented by other authors [48], [49] for these same species. It should be noted that the growth kinetics parameters are influenced by the culture conditions and the composition of the medium and reveal the good metabolic adaptation of the strains to the substrate [50].
Pediococcus doubling times have been reported to range between 1.60 and 5.80 h [6], values that are higher than those determined in this study (Table 2). In lactobacilli, the doubling time was between 1.37 and 2.69 h and the µx varied between 0.26 and 0.50 h-1, coinciding with other works that have reported similar kinetic parameters with doubling times between 1.21 and 2.56 h and specific growth rates between 0.270 - 0.573 h-1 for different Lactobacillus strains in selective culture media such as MRS [51]. For Lactococcus lactis, the doubling time was 1.83 h (Table 2), lower than that reported by Valencia-García et al. [21], who studied the kinetic parameters of LAB isolated from a Colombian fermented product. However, doubling times of 45 min and 1.1 h have been found in growth curves performed on selective media such as M17 [52]. Among the enterococci, E. faecium (24) presented the shortest doubling time and the highest specific growth rate (Table 2).
The kinetic parameter µmax (maximum specific growth rate) calculated by the logistic model, reflected significant differences between the strains (p<0.05), with P. acidilactici (21), E. faecium (24), W. viridescens (28), and L. citreum (20), presenting the highest µmax (Table 2). The values of µmax obtained in our study for these strains were similar when compared to other studies carried out with LAB in substrates such as Skim Milk and in selective culture media [30], [53]. Species of the genus Weissella have been highlighted in other investigations for presenting high growth rates, even higher than those of Leuconostoc [49] ; which coincides with the behavior observed in the present study (Table 2).
For the Lactobacillus genus, the values of the maximum growth rate were between 0.15 and 0.37 h-1 (Table 2), coinciding with what was reported for this genus when using whey as a substrate for lactobacilli fermentation [8]. Other authors, when evaluating the growth of L. fermentum in substrates such as soy milk, obtained values of µmax and doubling time lower than those reported in our study [54], with culture conditions and the composition of the fermentation medium being factors influencing the parameters of growth kinetics [50].
Validation is one of the steps necessary to verify that a model, whether experimental or simulated, has a satisfactory range of precision; in addition, it allows corroboration that the experimental data are consistent. The variations in the experimental and predicted biomass, using the logistic model, are shown in Figure 2, where the solid line refers to the data calculated with the logistic model and represents the adequacy of the mathematical model in describing LAB growth during growth kinetics in UHT milk. The results showed that for all strains, the R2 values in both models (experimental and logistic) were higher than the threshold of 0.8, which has been indicated as a good fit value [9]. In the LAB growth kinetics conducted in this work, the growth rate was proportional to the cell biomass, which agrees with the assumptions of the logistic model, indicating that the experimental data can be described according to this model.
The kinetic evaluation of the growth of the microorganisms to be used as starter cultures is necessary to optimize their industrial production [48], not only to assure a high cell density but to evaluate the different metabolites that can be produced in each of the growth phases such as organic acids, amino acids, fatty acids, exopolysaccharides, vitamins, acetoin, acetaldehyde, mannitol, bacteriocins and antioxidants [6]. In the results of this study, the strains belonging to the genera Lactococcus, Pediococcus, and Leuconostoc quickly adapted to milk, showing their potential to be used in cheese production. Lactococcus are considered as fast acidifiers leading to coagulation in a short time and producing flavors and texture during ripening [55].
Pediococcus is used in the production of fermented foods due to their tolerance to oxygen, growth at different pH levels, NaCl conditions, and temperature, characteristics that are required in the cheesemaking process. They are being studied to be applied in the modern food processing industry by their ability to produce bacteriocins and antimicrobial metabolites [56]. Leuconostoc plays important in the fermentation by its ability to preserve the product by acidification and contributing to the flavor and texture. Additionally, this specie can increase the nutritional quality of food by increasing digestibility [25]. P. acidilactici (21), E. faecium (24), Leu. mesenteroides (22) and W. viridescens (28) comprised the LAB group with the highest acidification potential, a parameter indicated as fundamental in curd formation and which, according to the literature [8], [21], becomes a selection criterion for the formulation of starter culture. Weissella has great potential in its application in food for its antagonistic activity against pathogens due to the production of several compounds like bacteriocins and organic acids. These strains can produce exopolysaccharides that can improve the texture of dairy products [57]. Even though Enterococcus has the ability to develop flavor and desirable aroma of dairy products, some strains are known to be opportunistic pathogens, produce biogenic amines, and transfer genes of virulence and antibiotic resistance [58]. Parameters that may be evaluated for their industrial use. Finally, it should be noted that the logistic model allowed for describing the behavior of the strains at an experimental level, a favorable situation at the time of conducting the industrial scaling up of these microorganisms. This study constitutes the initial stage in the formulation of a starter culture of autochthonous strains, which must be ratified through technological tests such as proteolytic and lipolytic activities, antimicrobial activity, NaCl and temperature tolerance, and production trials to determine their usefulness in the manufacture of double cream cheese with pasteurized milk.
4. CONCLUSIONS
P. acidilactici (21) and Leu. mesenteroides (22) had a rapid adaptation to UHT milk with lower pH values and high percentages of acidity, which shows their potential to be included in the native LAB starter culture. However, other strains such as W. viridescens (28) and E. faecium (24) that require more time to adapt to UHT milk were also able to reach acidity values lower than 5.1 in the same period (24 h). The logistic model was fitted to the experimental values of biomass for each strain. However, it is necessary to evaluate technological aptitude to validate the suitability of these strains as a starter culture.