Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Revista Colombiana de Ciencias Pecuarias
versão impressa ISSN 0120-0690versão On-line ISSN 2256-2958
Rev Colom Cienc Pecua v.19 n.1 Medellín jan./mar. 2006
ARTÍCULOS ORIGINALES
Polymorphism in 1908STR1934 locus of the 3’ UTR of the Nramp1 bovine gene in eight cattle breeds
Jenny P González S1, Microbiol y bioanalista; Omar A Saldarriaga C, MV, MSc1; Albeiro López-Herrera4, Zoot, MV, MSc, DrSci; Nelson R Bermúdez2, Zoot, MSc; Wildeman Zapata B1, Bact, Est MSc; Jorge E. Ossa L3, MV, MSc, PhD; María T. Rugeles L1, Bact, MSc, DrSci; Gabriel Bedoya B2, Biol, MSc.
1Grupo Inmunovirología-Biogénesis.
2Laboratorio Genética Molecular.
3Profesor Jubilado, Universidad de Antioquia, Apartado Aéreo 1226 Medellín Colombia.
4Grupo Biogem, Departamento de Producción Animal, Facultad de Ciencias Agropecuarias, Universidad Nacional de Colombia, Calle 59A No 63-20, Medellín- Colombia.
wzbuiles@virologia.udea.edu.co
(Recibido: 26 mayo, 2005; aceptado: 1 febrero, 2006)
Summary
The Nrampl gene has been associated with natural resistance to intracellular microorganisms in several species including bovine. Recent evidence suggests an association between polymorphism in the 3’ untranslated region (3’ UTR) of this gene with resistance/susceptibility (R/S) to Brucella abortus as determined in vivo and in vitro. In this study we tested for the variability of the short tandem repeat (STR) within the 3’ UTR of Nrampl in six breeds of Colombian creole cattle (CCC) and compared the genotypes with those of Holstein and Brahman, which were recently introduced into this country. In CCC as well as in Holstein we found the allele 175 fixed in all populations. In Brahman, 175 allele was also present with a frequency of 0.467 but additionally, in this breed there appeared five other alleles and among them two previously unreported: 183 y 185; also was found the allele 189 in the Colombian creole Harton del Valle cattle, which is not previously reported. Together these results suggest that the 175 allele in the 3’ UTR Nramp1 may be an ancestral allele in cattle and if this is true the association previously reported with the R/S trait requires further evaluation.
Keywords: colombian creole cattle, microsatellite, Nrampl, 3’ UTR,
Introduction
Resistance to infectious diseases is usually a complex genetic trait and recent data suggest that several resistance/susceptibility (R/S) associated genes are involved in more than one infectious disease (23). The gene Nramp coding for a Natural resistance-associated macrophage protein has been identified as the candidate gene controlling innate resistance to numerous intracellular pathogens such as Salmonella, Leishmania and Mycobacteria, in mice (21, 22). Nramp orthologous genes have been identified in several vertebrates species such as human (10, 22), chicken (8), swine (20), equine (7), and in ruminants like sheep, red deer (13), American bison (Feng et al., Texas A&M University, College Station, TX USA, unpublished), Water buffalo (Hashad et al. Texas A&M University, College Station, TX USA, unpublished) and bovine (6).
The Nramp gene expresses exclusively in the monocyte/macrophage lineage and encodes a phosphoglycoprotein (24) that belongs to an ancient family of membrane proteins (5). Recent evidence supports the idea that one of its roles in mouse macrophages resistant to Mycobacterium bovis is to transport metal ions such as iron into the bacteria containing phagosome where it serves as a catalyst of the Fenton/Haber-Weiss reaction to increase the capacity of the cell to limit bacterial growth (9; 18).
Feng et al 1996, cloned the cDNA bovine homologous of the mouse Nramp; the gene was mapped on BTA 2 (chromosome 2), syntenic loci conserved in mice and humans and it is known that it encodes for a protein of 548 residues.
Horin et al 1999; found a complex microsatellite pattern within the 3’ untranslated region (3’ UTR) of the Nramp1 bovine gene. When they compared the original sequence of Feng with the sequence found in unrelated cattle there appeared a variation short tandem repeat (STR) GT and a transversion T®G in the 1782 position of the 3’ UTR. The original sequence has a 1782/T + (GT)11 = (GT)12 and two of the clones obtained by Horin (7) have 1782/G + (GT)10.= (GT)10.
This investigator proposed the designation Nramp1.1 for Feng’s sequence and Nramp 1.2 for the new allele.
The functional significance of this new allele in resistance/susceptibility is still unclear. However, preliminary studies have shown that polyphormism, based on single stranded conformational analysis (SSCP) in the 3’ UTR of bovine Nramp1 presented a significant association (p=0.0089) with the phenotype of resistance/susceptibility in 22 unrelated animals, previously phenotyped using an in vivo challenge of virulent Brucella abortus (6). The different patterns in SSCP are due to the size differences in the 3’UTR STRs and so far only one of four alleles (175) has been associated with resistance to B. abortus (Adams et al, Texas A&M University, College Station, TX USA, Personal communication).
Association of Nramp with resistance/susceptibility was first described in mice where it was found that a transition 783 G®A producing a change from G169D in the Nramp protein transforms the phenotype to M. bovis from resistance to susceptibility (22). In humans, polymorphism in the 3’ UTR has also been associated with susceptibility to tuberculosis (2).
Colombian Criollo Cattle (CCC) is derived from the animals brought by Christobal Columbus in his second trip and there are seven distinct breeds: Blanco Orejinegro (BON), Chino Santanderiano (ChS), Sanmartinero (SM), Costeño con Cuernos (CC), Hartón del Valle (HV), Romosinuano (RS) and Casanareño (CS). These breds have survived for over 500 years, in spite of the conditions of the tropics together with its parasites and also the socioeconomic conditions which include an extensive type of management, with no selection and the presure of foreign breeds, specially Brahman and Holstein arriving in this country during the last 150 years.
BON in particular still has a high genetic variability comparable to African bovine populations (4). This BON breed characterizes itself by its adaptability to diverse ecological conditions, such as those found in the tropic; including tolerance to heat and to internal and external parasites, ability for grazing and browsing and a remarkable capacity to use low-quality forages. Additionally, this breed has shown high longevity and good fertility (12). This work belongs to a research line whose main goal is to characterize the CCC to obtain good arguments for its rescue from the endangered condition in which they are currently classified; specifically, we studied the STR variability within the 3’ UTR of the Nramp gene to further look for association of some alleles in this microsatellite locus with R/S to intracellular pathogens such as Brucella.
Materials and methods
Bovine populations
Samples were collected from 131 bovine representing 3 different pure breeds, BON (Bos taurus), Holstein (Bos taurus) and Brahman (Bos indicus). Additionally we included 181 samples from five other CCC breeds (HV, SM, CC, RS, ChS), average 36 per breed. A description of the sample is presented in table 1.
DNA was extracted with the phenol chloroform technique (17) from peripheral blood. The segment (nucleotide positions 1813-1988) which contains an GT repeat array in the 3’ UTR of Nramp gene, was amplified by PCR using the following primers (FW 5’- AAGGCAGCAAGACAGACAGG-3’ and RV 5’- ATGGAACTCACGTTGGCTG-3’) this sequences were kindly provided for Dr. G. Adams, (Texas A&M University, College Station, TX USA). 200 ng of DNA were amplified in a volume of 25 ul as follows: 7.5 pmol of each primer, 0.3 units of Taq DNA (PROMEGA, Madison, USA), 1mM of MgCl2 and 150 uM of each dNTP. These samples were incubated at 95°C/ 2 min, and then 30 cycles of 94°C /45 sec, 63°C /30sec, 72°C/ 60 sec, and finally 72°C/ 7min. For this procedure we used a 2400 GeneAmp PCR system (Perkin Elmer, Norwalk CT, USA). Additionally, in order to compare the products between the 175/175 BON and Holstein we amplify another fragment among the nucleotides 1697 up to 2203 using the following primers (FW 5’-GAGAAGGGGAGGACCTCGGG-3’and RV 5’- CAGTTTGGAGCCTGGGTCCC-3’) published previously by Feng et al (6), and the same reaction conditions as described above.The PCR products were confirmed in a 2% agarose gel electrophoresis and stained with ethidium bromide.
Single-strand conformation polymorphism analysis (SSCP)
SSCP of the PCR-products was carried out as follows: The PCR products were denatured (90°C/ 2min) and subjected to electrophoresis in a 6% nondenaturating polyacrylamide gel. The migration patterns were resolved using silver nitrate (Sigma, St. louis, USA) and the samples were compared (14).
Allelic size determination
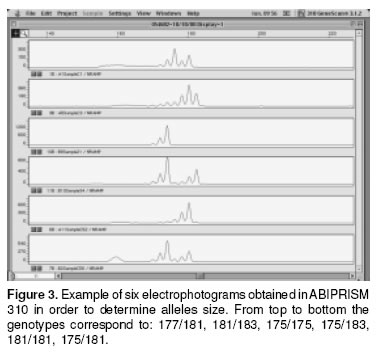
The size of the allele was determined only in those samples exhibiting a different SSCP migration pattern, with reference to a control homozygous sample of known size (175 pb) previously characterized by Adams G. et al, Texas A&M University, College Station, TX USA; (personal communication) using a genetic analyzer ABIPRISM 310 (Applied Biosistems, Foster City, CA), with the GENESCAN and GENOTYPER software.
DNA sequencing
In order to search for possible differences between a homozygous 175 BON allele and homozygous 175 Holstein allele, the sequence of a segment (nucleotide positions 1697-2203, 3’ UTR) of one animal (175/175 genotype) belonging to each breed was determined. The sequence reactions were carried out with the DNA sequencing kit Big Dye terminator (PerkinElmer, Foster City, CA), following the manufacturer’s recommendations for template and primer concentrations and cycling conditions. The products were resolved in a genetic analyzer ABIPRISM 310 (Applied Biosistems, Foster City, CA), analyzed with the navigator software.
Analysis of population genetic structure
In order to assess the allele neutrality and population independence, two Wright’s Statistics (Fis, Fit), and the GENEPOP software were used.
Results
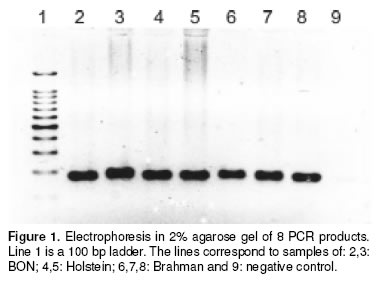
Although some PCR products exhibited slightly different sizes we could not resolved the fragments with the 2% agarose gel (see Figure 1).
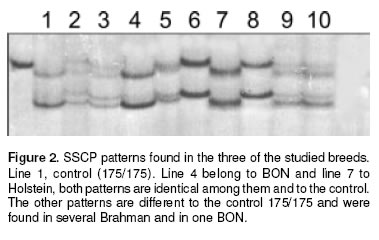
The detected electrophoretic SSCP patterns in BON, Brahman and Holstein are shown in figure 2. All Holstein and all but less one BON had the 175/175 pattern, this animal showed 175/183 genotype; while 20 of 76 Brahman also had 175. On the other hand all the other 5 CCC breeds studied presented 175/175 Genotype in most of the samples; with the only exception of 2 of HV samples which exhibited 175/ 177 and 175/181, two of the ChS samples (175/181 genotype), two of the CC samples (175/181 genotype) and four of the RS samples (Three of them 175/181 genotype and the other 181/181) (Data not shown).
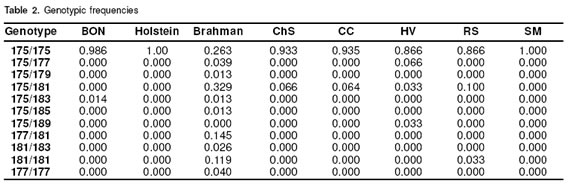
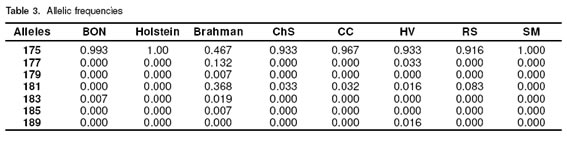
In figure 3 some electrophotograms obtained with the ABIPRISM 310 are shown. The picks correspond to different allele sizes. In table 2 the results of genotyping of the eigth cattle breeds can be seen. It was confirmed that the SSCP pattern really has a 175 allele in a homozygous condition. All the samples from Holstein cattle used in this study showed 175/175 genotype (genotypic frequency 1.00), the BON animal with a different pattern corresponded to 175/183 genotype (genotypic frequency 0.014). In the Brahman samples not having 175, five different size alelles (177,179, 181, 183, 185) were found. In this breed three homozygous genotypes were detected: 175/175, 177/177, 181/181 with a frequency of 0.263, 0.039, 0.119, respectively. Of the Brahman samples analyzed 0.578 were in heterozygous conditions, from which 0.407 shared the 175 allele. Additionally in the Brahman breed there were two alleles (183, 185) no previously reported. In the other CCC breeds also the 175/175 genotype was the most frequent, and this phenotype is the only founding the SM samples; the HV breed was the most variable in the genotypes and one sample showed the phenotype 175/189, and the allele 189 was not previously reported.
With respect to allelic frequencies the most prevalent was 175 in all the breeds. In all the breeds except Brahman this allele is fixed, in Holstein and SM this allele was in a frequency of 1.000, and in Brahman the frequency was near 50%. The 179, 185 and 189 alleles were the less prevalent in all the breeds (see Table 3).
Since in BON and Holstein there was no genetic variability the Fis value was calculated only for Brahman to test for Hardy-Weinberg (H&W) equilibrium. The Fis value in Brahman was 0.085 and the P value for heterozygous deficit was 0.0942; it’s means that this population is in H&W equilibrium, for the 3’ UTR- STR locus.
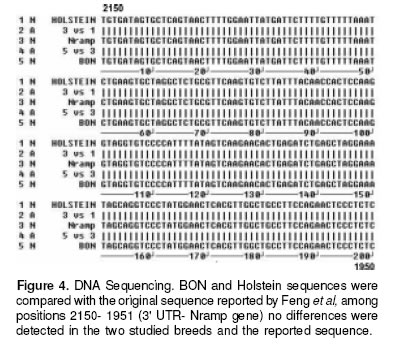
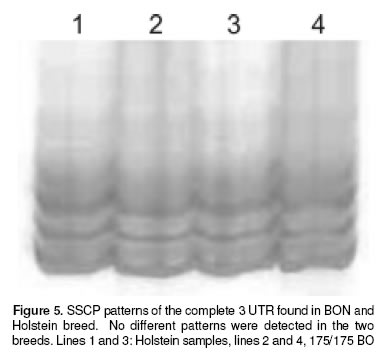
Finally, in the figure 4 is showed the sequence for the STR of one Holstein an one bon samples, compared with a sequence reported for this STR by Feng et al (6 ), the sequence of our samples were identical to the reported sequence. We also performed a SSCP of the complete 3´-UTR Nramp1 looking for a complex pattern of the microsatellite polymorphism reported by Horin et al (7), but no differences were found in this technique (see figure 5).
Discussion
We found a good correlation ship between the SSCP pattern and the allele size obtained by ABI-PRISM, so that those samples with similar SSCP patterns have the same genotype while those samples with different SSCP patterns exhibit different genotypes. Therefore the SSCP technique is appropriate to determine variability in the 3’UTR- STR of the Nramp1 gene. Consequently we decided to genotype other five CCC breeds with this method and we found the 175 allele fixed in all them as in BON and Holstein.
The finding of the 175 allele fixed in seven of the eight studied breeds (including BON and Holstein) makes us believe that this phenomenon is the effect of selection, i.e. it is associated to some kind of resistance to intracellular infectious agents, since polymorphism in 3’-UTR of Nramp1 have been associated with mRNA stability in other species including humans (2). Additionally, a preliminary study has found association (P= 0.0089) (6) of the 175 allele (Adams et al, personal communication) with phenotypic resistance to Brucella abortus infection as determined in vivo (19) and in vitro (15, 16).
Since the results of identical genotype for BON and Holstein were somehow against our intuition that BON shall be more resistant than Holstein we tested for the possibility that the up and downstream regions off 1908(GC)131934 3´-UTR Nramp1 locus, the object of this work, could be different for these breeds. But mainly we wanted to emphasize the 1781(GC)121804 3’-UTR Nramp1 locus where Horin et al had found a complex polymorphism pattern in cattle. This would eventually tell us if in spite of these breeds having the same size STR allele they would have polymorphism outside the target locus; in which case the supposed resistance of BON could be explained by a hitch-hiking phenomenon. As can be seen in figures 4 and 5, no difference was found neither with the SSCP technique for the whole 3’ UTR nor with the sequencing procedure in the downstream region.
The finding of identity up and downstream of 175 in both BON and in Holstein does not mean that this allele is not associated with R/S. It would be necessary to determine with Holstein or eventually the familly of Holstein tested are indeed resistant to Brucella. It was at this point when we judged important to test the genotype of the other 5 CCC breeds where again the allele was fixed (169 samples out of 171). The association of this genotype with R/S in CCC could not be understood until the experimental challenge is made. However, these intriguing results deserve at least an evolutive nterpretation.
The monomorphism found in the CCC and Holstein breeds for the locus 1908STR1934 3´-UTR Nramp1 could be due to inbreeding and/or genetic drift as an effect of husbandry; but this idea it is not in full agreement with the results of Carvajal et al 1998 and Bedoya et al 2001, whose works with five microsatellite markers in CCC indicated that heterocigocity in these cattle populations was very high (0.672); interestingly, however, in their studies they also reported a H&W disequilibrium (P= 0.0006). This last finding does agree with our results.
A second scenario refers to the possibility that the fixation of the allele 175 in these breeds is due to positive ancestral selection as an adaptative trait (intracellular parasites, for instance). However, in Brahman, not withstanding the higher frequency of 175 (0.467), other five alleles were found, and the population was in H&W equilibrium (Fis=-0.085 and P= 0.0942) for the allelic frequencies of the 1908STR1934 locus. This means that the polymorphism in the studied locus in Brahman and perhaps in cattle in general is neutral. The last possibility is that the allele 175 is ancestral appearing, in taurina after the coalescence between B. taurus and B. indicus since it was monomorphic in CCC and in Holstein (both of them B. taurus). The highest frequenccy of the 175 allele in Brahman can be explained by the fact that this breed is a blend of indicus and Taurus (11).
It has been clearly demonstrated in mice that the Nramp gene has indeed a broad pleiotropic effect on macrophage activation pathways which speaks of the complexity of the role of the gene in the R/S phenomenon (Reviewed by 3). Our results are applicable only to one locus within that gene.
From this point we think that research on this field should be directed toward the identification of polymorphism in the coding and in the promoter regions of the bovine Nramp1 gene to further associate eventual genotypes with phenotypic R/S traits tested by in vivo and in vitro challenges. Additionally it would be interesting to search for polymorphism in other genes with which Nramp has been reported to interact, such as iNOS, CXC Chimoquines, TNFa, Bola Class II, and others (3). Alleles of the genes coding for these proteins could be found in linkage desequilibrium with alleles of Nramp by a natural selection processes.
Finally we shall conclude with the major lesson that we have reaffirmed with the outcome of our work: knowing as we know now that R/S traits are complex and that Nramp is pleiotropic it does not make sense to look deterministically on a single locus within one single gene, because what we are bound to find is that polymorphism in a given locus seems to be neutral, as we suspect out of our results.
Acknowledgments
This study was supported by grant from Committee for the Development of the Investigation of the University of Antioquia, project number, CODI Number R118.355.01
Resumen
Análisis de polimorfismos en el locus 1908STR1934 locus del 3’ UTR del gen Nramp1 bovino en ocho razas de ganado
El gen Nramp1 ha sido asociado con resistencia natural a microorganismos intracelulares en varias especies incluyendo bovinos. Evidencia reciente sugiere una asociación in vivo e in vitro, entre el polimorfismo en la región 3’ no traducida (3’ UTR del inglés 3’ untranslated region) de este gen con resistencia susceptibilidad (R/S) a Brucella abortus. En este estudio se evaluó la variabilidad de una secuencia repetida en tandem (STR, del inglés short tandem repeat) dentro del 3’ UTR de Nramp1 en seis razas de ganado criollo colombiano (GCC) y se comparó con la variabilidad de los de ese STR en Holstein y Brahman, razas que fueron recientemente introducidas a este país. En GCC así como en Holstein se encontró que el alelo 175 esta fijado en todas las poblaciones. En Brahman. El alelo 175 estuvo presente con una frecuencia de 0.467, pero adicionalmente en este ganado aparecieron otros 5 alelos, dos de ellos no habían sido previamente reportados: 183 y 185; también se encontró el alelo 189 en el ganado criollo colombiano Harton del Valle, el cual no está reportado previamente. Estos resultados sugieren que el alelo 175 en el 3’ UTR de Nramp1 puede ser un alelo ancestral en ganado y si esto es cierto, la asociación previamente reportada con la característica de R/S requiere evaluación futura.
Palabras clave: ganado criollo colombiano, microsatélite, Nramp1, 3’ UTR,
References
1. Bedoya G, Carvajal LG, Bermúdez NR, Moreno FL, Márquez ME, et al. Estructura molecular y poblacional del ganado criollo Colombiano. Rev Col Cienc Pec. 2001; 14: 109-120. [ Links ]
2. Bellamy R, Rawende C, Corrah T, McAdam KPWJ, Whitte HC, et al. Variations in the Nramp1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998; 338: 640-644. [ Links ]
3. Blackwell JM, Searle S. Genetic regulation of macrophage activation: understanding the function of Nramp1 (=Ity / Lsh / Bcg). Immunol Lett. 1999; 65: 73-80. [ Links ]
4. Carvajal LG, Bermúdez NR, Moreno LF, Márquez ME, Estrada JL, et al. Genetic diversity of colombian criollo cattle. Anim Genet. 1998; 29: 41.47. [ Links ]
5. Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, et al. Nramp1 defines a family of membrane proteins. PNAS. 1995; 92: 10089-10093. [ Links ]
6. Feng J, Li Y, Hashad M, Schurr E, Gros P, et al. W. Bovine natural resistance associated macrophage protein 1 (Nramp1) gene. Genome Res. 1996; 6: 956-964. [ Links ]
7. Horin P, Rychlik L, Templeton JW, Adams LG. A complex pattern of microsatellite polymorphism within the bovine Nramp1 gene. Eur J Immunogenet. 1999; 26: 311-313. [ Links ]
8. Hu J, Bumstead N, Barrow P, Sebastiani G, Olien L, et al. Resistance to salmonellosis in the chicken is linked to Nramp1 and TNC. Genome Res. 1997; 7: 693-704. [ Links ]
9. Kuhn DE, Baker BD, Lafuse WP, Zwilling BS. Differential iron transport into phagosomes isolated from the RAW264.7 macrophage cell lines transfected with Nramp1Gly169 or Nramp1Asp169. J. Leukoc. Biol. 1999; 66:113-119. [ Links ]
10. Liu J, Fujiwara M, Buu NT, Sánchez FO, Cellier M, et al. Identification of polymorphisms and sequence variants in the human homologue of the mouse natural resistanceassociated macrophague protein gene. Am J Hum Genet. 1995; 56: 845-853. [ Links ]
11. Machugh DE, Shriver MD, Loftus RT, Cunningham P, Bradley DG. Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle. (Bos taurus and Bos indicus). Genetics.1997; 146:1071-1086. [ Links ]
12. Martínez G. The Colombian Criollo Cattle Breeds. In: Proceedings of the third global conference on conservation of domestic animal genetic resources. Rare breeds international; Queens University, Kignston, Ontario- Canada. 1995; 161-166. [ Links ]
13. Mathews GD, Crawford AM. Cloning, sequencing and linkage mapping of the Nramp1 gene of sheep and deer. Anim Genet. 1998; 29: 1-6. [ Links ]
14. Orita M, Suzuki Y, Seliya T, Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphims using the polymerase chain reaction. Genomics. 1989; 5:874-879. [ Links ]
15. Price RE, Templeton JW, Smith III R, Adams LG. Ability of Moncuclear phagocytes from cattle naturally resistant or susceptible to brucellosis to control in vitro intracelullar survival of Brucella abortus. Infect Immun. 1990; 58: 879-886. [ Links ]
16. Qureshi T, Templeton JW, Adams LG. Intracellular survival of Brucella abortus, Mycobacterium bovis- BCG, Salmonella dublin, and Salmonella typhimurium in macrophages from cattle genetically resistant to Brucella abortus. Vet. Immun. Immunopat. 1996; 50: 55-65. [ Links ]
17. Sambrook J, Fritsch EF, Maniatis T. Isolation of highmolecular- weight DNA from mammalian cells. In: Sambrook J, Fritsch EF, Maniatis T., editors. Molecular cloning. A laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. p. 9.14-9.23. [ Links ]
18. Swilling BS, Kuhn DE, Wikoff L, Brown D, Lafuse W. Role of iron in Nramp 1- mediated inhibition of mycobacterial growth. Infec Immun. 1999; 67: 1386-1392. [ Links ]
19. Templeton JW, Adams L.G. Natural resistance to brucellosis: in Advances in Brucellosis Research. Garry Adams ed., 1990: Cap 10. [ Links ]
20. Tuggle CK, Schmitz CB and Gingerich-Feil D. Cloning of a pig Full-length natural resistance associated macrophage protein (Nramp1) cDNA. J. Anim. Sci. 1997, 75: 277. [ Links ]
21. Vidal SM, Gross P. Resistance to infection with intracellular parasites: Identification of a candidate gene. NIPS. 1994; 9: 178-183. [ Links ]
22. Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural Resistance to infection with intracellular parasites: Isolation of a candidate candidate for BCG. Cell. 1993; 73: 469-485. [ Links ]
23. Vidal SM, Tremblay ML, Govoni G, Gauthier S, Sebastiani G, et al. The Ity/Lsh/Bcg locus: Natural resistance to infection with intracellular parasites is abrogated by disruption of the of the Nramp1 gene. J Exp Med. 1995; 182: 655-666. [ Links ]
24. Vidal SM, Pinner E, Lepage P, Gauthier S, Gros P. Natural resistance to intracellular infections: Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1 D169) mouse strains. J Immunol. 1996; 157: 3559-3568. [ Links ]