Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Revista MVZ Córdoba
versão impressa ISSN 0122-0268versão On-line ISSN 1909-0544
Rev.MVZ Cordoba vol.24 no.3 Córdoba set./dez. 2019 Epub 04-Jun-2020
https://doi.org/10.21897/rmvz.1232
Research article
Incidence of Escherichia coli 0157:H7 in feces of lactating ruminants with diarrheal syndrome
1 Universidad Autónoma de Zacatecas, Unidad Académica de Medicina Veterinaria y Zootecnia, Km 31.5 de la carretera panamericana tramo Zacatecas-Fresnillo, Enrique Estrada, Zacatecas, México.
2 Universidad Autónoma de Zacatecas, Unidad Académica de Ciencias Biológicas, Avenida preparatoria s/n colonia Hidráulica, Zacatecas, Zacatecas, México.
3 CONACYT-Consorcio CENTROMET, Camino a Los Olvera #44, Corregidora, Querétaro, México.
Objective.
To identify Escherichia coli 0157:H7 present in diarrheal feces of lactating ruminants with diarrheal syndrome and safety of colostrum intake.
Materials and methods.
A feces sampling of 316 ruminants was carried out during the period of August 2015 to March 2016 in the municipalities of Río Grande, General Enrique Estrada, Morelos and Calera de Victor Rosales of the state of Zacatecas, obtained from 67 cattle, 183 sheep and 66 goats.
Results.
The following were identified in CHROMagarTM chromogenic medium: 260 coliforms, 78 Escherichia coli 0157:H7, 16 Proteus spp. and 25 colonies of unidentified bacteria, finding an incidence of Escherichia coli 0157:H7 of 22.03% in the four municipalities.
Conclusions.
Escherichia coli 0157:H7 is the second bacteria found in ruminant feces with an incidence of 22%, which is a mortality risk factor in lactating ruminants (less than 21 days old), causing economic loss and health risk for the population of the state of Zacatecas.
Keywords: Bacteria; chromogenic medium; sheep (Source: CAB)
Objetivo.
Identificar Escherichia coli O157:H7 presente en heces diarreicas de rumiantes lactantes con síndrome diarreico y seguridad de ingesta de calostro.
Materiales y métodos.
Se realizó un muestreo de 316 rumiantes durante el período de agosto 2015 a marzo 2016 en los municipios de Río Grande, General Enrique Estrada, Morelos y Calera de Víctor Rosales del estado de Zacatecas, 67 de bovinos, 183 de ovinos y 66 de caprinos.
Resultados.
Se identificaron en medio cromogénico CHROMagarTM: 260 coliformes, 78 Escherichia coli O157:H7, 16 Proteus spp., y 25 colonias de bacterias sin identificar con este medio, encontrándose una incidencia de Escherichia coli O157:H7 de 22.03% en los cuatro municipios.
Conclusiones.
Escherichia coli O157:H7 es la segunda bacteria encontrada en heces de rumiantes con un 22% de incidencia, la cual es un factor de riesgo de muerte en rumiantes lactantes (menos de 21 días de nacidos) causando pérdidas económicas y riesgo para la salud de la población del estado de Zacatecas.
Palabras clave: Bacterias; medio cromogénico; ovinos (Fuente: CAB)
INTRODUCTION
The economic repercussion resulting from digestive diseases in ruminants in Mexico is serious and frequent. The pathological symptoms presented by the ruminants under this type of stress are as follows: low feed consumption, presence of diarrheas (or dysentery), ruminal acidosis, laminitis, limp, delay in corporal development of lambs, kids and calves 1. This means an unforeseen added cost for the producer, along with expenses in veterinary treatments 2.
In intensive production systems, the mortality index of lambs due to digestive diseases prior to weaning tends to be less than 10%, whereas in extensive systems the losses can be as high as 53% of the lambs born 3. However, various reports mention that infectious processes and nutritional and metabolic disorders represent the principal cause of neonatal mortality 4. In fact, between 15 and 20% of the neonatal losses can be attributed to digestive diseases, and consequently, the rest (around 80%) are the result of environmental or management factors 3.
The Centers for Disease Control (CDC) of the Department of Health and Human Services of the United States mention that Escherichia coli is a type of bacteria that normally lives in the intestines of humans and animals. For the most part they are inoffensive and in fact are part of a healthy intestinal tract. However, some serotypes are pathogens, causing diarrheas or diseases outside of the intestinal tract 5.
Escherichia coli is classified into six serotypes that are associated with diarrhea; among these is enterohemorrhagic Escherichia coli (EHEC), which causes hemorrhagic colitis (HC) and is identified through the production of Shiga toxins (Stxs). Shiga toxin-producing Escherichia coli (STEC) is a group of zoonotic pathogens, and Escherichia coli 0157:H7 is the serotype associated with hemorrhagic colitis and hemolytic uremic syndrome (HUS), principally in the United States.
Laven et al 7 demonstrated that the presence of Escherichia coli in ruminants was higher in the colon than in the rumen, associating the presence of Escherichia coli to the ingestion phase and not to the intestinal wall of the rumen. Studies have demonstrated that the terminal part of the colon is the principal development site of the colonization of Escherichia coli 0157:H7 and that the lymphoid tissue located in the recto-anal junction is the colonization site in cattle 8. Therefore, this colonization is considered a “super-propagator” which causes severe infections with epidemiological manifestations characterized by hemorrhagic diarrheas, colitis and hemolytic uremic syndrome 9.
Adequate isolation and identification methods are crucial for the diagnosis of Escherichia coli 0157:H7. Before confirming its presence in a sample, it is necessary to isolate and identify it in selective and differential culture media. The media used conventionally for the isolation of Escherichia coli 0157:H7 include Sorbitol MacConkey Agar (SMAC), CefiximeTellurite Sorbitol MacConkey Agar (CT-SMAC), CHROMagarTM 0157 (CHROMagar), Tellurite CHROMagarTM 0157 (T-CHROMagar) and Vancomycin Cefixime Cefsoludin CHROMagarTM 0157 (VCCCHROMagar) 6,10.
The chromogenic chemical substrates patented for the identification of microbial colonies led to the development of a wide range of media for the detection of pathogens. The medium CHROMagarTM 0157 selectively promotes the growth of Escherichia coli 0157:H7 and the difference from Escherichia coli no. 0157, due to a chromogenic substrate of β-galactosidase and a change in the pH 6. This medium is used for the detection of Escherichia coli 0157:H7 in alimentary as well as environmental samples, but could be more selective with the addition of potassium tellurite, or with vancomycin, cefixime and cefsoludin 11.
Therefore, the objective of the present study was to identify Escherichia coli 0157:H7 in diarrheic feces of lactating ruminants younger than 21 days through the use of agar CHROMagarTM, which will permit producers to make decisions about which treatments to employ in order to avoid the incidence of this pathogen and reduce neonatal mortality.
MATERIALS AND METHODS
Samples. 316 swabbed rectal samples from 183 sheep, 66 of goats and 67 of cattle under 21 days of age were obtained, and with the security of having ingested colostrum, with presence of diarrheic syndrome. The samples were collected rectally with a sterile cotton swab, and were labeled and transported in Stuar® medium. The sampling was carried out during the period of August 2015 to March 2016 in four municipalities of the state of Zacatecas: in Calera de Victor Rosales, the localities El Bordo, Noria de Agostadero, El Duraznillo, El Vergel and El Porvenir: in Morelos the localities of Hacienda Nueva, Noria de Gringos, Las Pilas and El Palmar: in General Enrique Estrada, Los Ángeles, Puentecillos and San Sebastián, and in Río Grande the localities El Carrizal, Los Ramírez, San José de Ranchitos and Tierra Blanca (Figure 1). (Coordinates: El Bordo 102°49’20”O 22°57’02”N, San José de Ranchitos 102°57’35”O 23°40’0”N, El Carrizal 103°01’08”O 23°38’25”N, El Duraznito 102°38’26”O 23°01’29”N, El Palmar 102°39’58”O 22°53’24”N, El Porvenir 102°40’00”O 22°56’42”N, El Vergel 102°41’01”O 22°59’58”N, General Enrique Estrada 102°44’29”O 22°59’40”N, Hacienda Nueva 102°36’21”O 22°49’30”N, Las Pilas 102°36’46”O 22°50’24”N, Los Ramírez 103°01’52”O 23°50’49N”, Los Ángeles 102°43’22”O 22°58’43”N, Noria De Agostadero 102°46’59”O 22°53’14”N, Noria de los Gringos 102°43’04”O 22°50’53”N, Puentecillos 102°45’03”O 23°02’47”N, San Sebastián 102°47’54”O 23°03’37”N, Tierra Blanca 103°04’36”O 23°49’38”N).
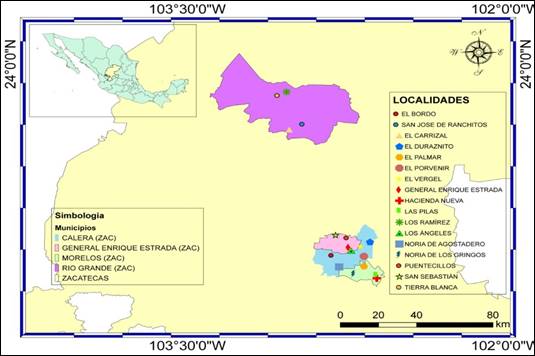
Figure 1 Geographic location of the localities sampled in the four municipalities of the state of Zacatecas.
Gram negative bacterial culture. The cultures were made in duplicate. Each sample was sown in Petri jars with MacConkey agar (SMAC), and incubated in a bacteriological oven at 37 °C for 48 hours 12. The creamy pink colonies were considered positive and were individually isolated for their later re-sowing.
Identification of bacteria in chromogenic medium CHROMagarTM. After the bacteria were isolated in MacConkey agar, a colony was taken for the identification of the serotype of Escherichia coli 0157:H7 or its non-mobile variant 0157:H (serotype VTEC), which is more common in public health. The re-sowing was made by plate striation in chromogenic medium CHROMagarTM and was incubated at 37 °C during 24 hours 13 (Table 1).
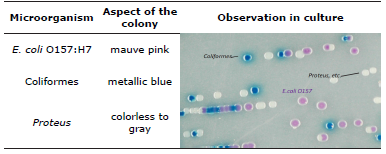
The colonies of Escherichia coli 0157:H7 grow in CHROMagarTM 0157 and produce a mauve pink color due to chromogenic substrates in the medium, thus permitting the presumptive identification of the primary isolation plate and the differentiation from other organisms 13 (Table 1).
Statistical analysis. The experimental data were analyzed first with descriptive statistics, and afterwards using a classification table or contingency table. The Chi-squared statistical analysis was used, which is a statistic that makes it possible to determine the degree of independence among the variables. The assumption of this test is as follows: Ho: The prevalence of the bacteria is independent from the type of ruminant the carrier may be; Ha: The prevalence of the bacteria is dependent on the type of ruminant carrier. The most important index of this test is the value p. In general, when the value p is less than 0.05, Ho is rejected and it is concluded that the bacteria are not independent. Al of the statistical process was carried out with SPSS 25.0 for Microsoft Windows.
RESULTS
The determination of Escherichia coli 0157:H7 was made using chromogenic medium CHROMagarTM differentiating the bacterial colonies by color; Escherichia coli 0157:H7 presented mauve pink coloration, Proteus ssp. are from colorless to gray and the coliforms present metallic blue colored colonies, as can be seen in Figure 2.
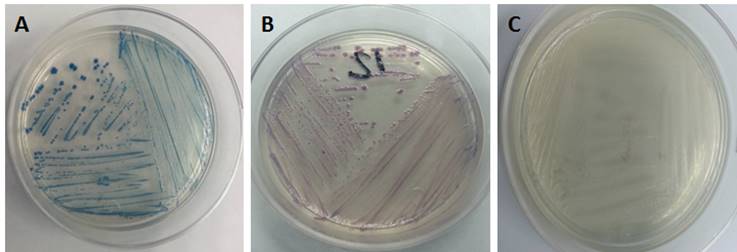
Figure 2 Development of bacteria in CHROMagarTM. A) Coliforms, B) E. coli O157: H7 and C) Proteus spp.
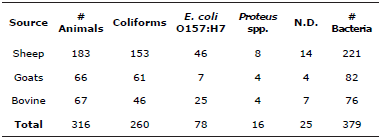
Some samples presented development of two or more colonies with distinct morphology and coloration, thus the total number of the bacterial colonies identified was 354, and in 25 cases there was no development in 24 hours, despite the fact that development was found in MacConkey agar and colonies with descriptive morphology for Escherichia coli there was no development in CHROMagarTM.
260 coliforms were identified, along with 78 Escherichia coli 0157:H7 and 16 Proteus spp., in all of the ruminants sampled, of which 183 were sheep, where it was found that in 46 sheep there was presence of Escherichia coli 0157:H7, in goats it was found in 7 of 66 and in cattle, 25 of 67 as is shown in Table 2. Furthermore, using the chi-squared test it is confirmed that the prevalence of the bacteria (coliforms, Escherichia coli 0157:H7 and Proteus spp.) depends on the type of ruminant (sheep, cattle and goat) which is the carrier of the bacteria (Table 3).
Table 3 Contingency table: type of bacteria vs ruminant type.
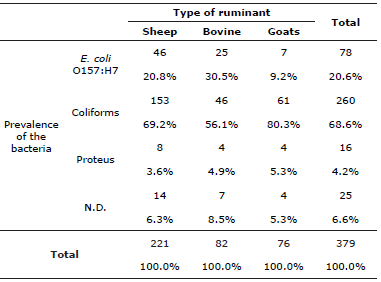
Ho: The prevalence of bacteria is independent of the type of ruminant that is the carrier; Ha: The prevalence of bacteria is dependent on the type of ruminant that is the carrier. Chi-square value of Pearson=13.243. gl=6; p=0.039. As the value of p≤0.05 is rejected Ho. N.D.: No Determinados.
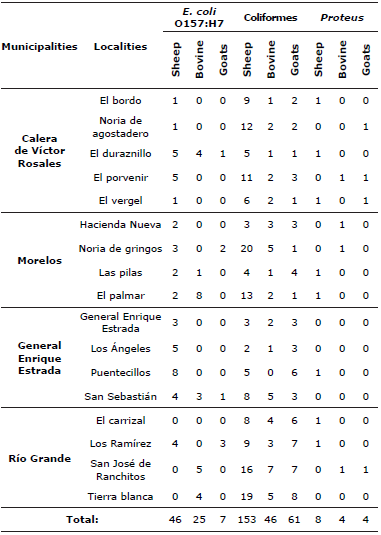
The distribution of bacteria found according to their geographic location correspond to the following: 18 Escherichia coli 0157:H7, 60 coliforms and 7 Proteus ssp., in the municipality of Calera de Victor Rosales; 40 Escherichia coli 0157:H7, 11 coliforms and 9 Proteus ssp., in localities of Morelos, 24 Escherichia coli 0157:H7, 41 coliforms and 1 Proteus ssp., in localities General Enrique Estrada and 16 Escherichia coli 0157:H7, 99 coliforms and 4 Proteus ssp., in localities of the municipality of Río Grande (Table 4).
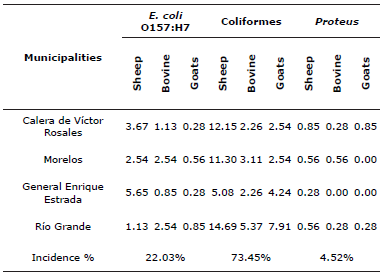
The highest incidence of bacteria were the coliforms with 73.45% in the four municipalities, followed by Escherichia coli 0157:H7 with 22.03% and of Proteus with 4.52% (Table 5). With these results, the producers of the region are able to make the best decision of prevention treatments to reduce the incidence of this pathogen and thus reduce mortality in lactating ruminants.
DISCUSSION
The presence of Escherichia coli 0157:H7 in cattle should be taken into consideration as a possible problem of public health, given that this bacteria strain is present in feces and is a source of infection for humans. In the finishing process of the animal, refrigeration of the carcass and also in milking, contamination of the meat and milk may occur 14.
It has been reported that the presence of Escherichia coli 0157:H7 in cattle feces seems to be influenced by the age of the animal 15. For example, calves under eight weeks of age (56 days) and heifers excrete more of the Escherichia coli strain than adult cattle 16.
Escherichia coli 0157:H7 has become an important problem for human health, principally in the United States 6; Olvera et al 16 have stated that cattle are the primary reservoir of the bacteria Escherichia coli 0157:H7, which causes hemolytic uremic syndrome in humans. In fact, the carcasses from feedlot systems present the highest incidence of verotoxigenic Escherichia coli associated with hemolytic uremic syndrome, with respect to the grazing system where vaccination reduced the contamination of these bacteria in beef by 54.1%.
To diagnose this pathogen, adequate isolation and identification methods are crucial. There is little information available on its prevalence in cow/calf management. However, in Louisiana, U.S., an 8% prevalence of this pathogen has been reported, without significant differences among fecal matter, water (from water troughs and ponds) and surface smears (of water troughs and hay bunk) as sources of contamination 6.
In the investigation conducted by Fox et al 18, a general prevalence of 62% was found, higher than what was obtained in the present study (22.03%). The prevalence of Escherichia coli 0157:H7 in cattle is variable among each individual and each herd. In stabled cattle, it can be found in a range of 10 to 28%, but it can be higher, as much as 80% in the summer months 19. Cattle are the most affected, followed by sheep 20, which indicates there is no host selection for Escherichia coli 0157:H7.
With the use of CHROMagarTM it has been possible to rapidly determine the incidence with an identification sensitivity of 98% and a specificity of 100% 13, being superior to MacConkey sorbitol agar, which despite having the same sensitivity, its specificity decreases to 85% and has a precision of 86%. This medium was initially used only in the food industry for the rapid release of pathogen free foods, but it has been tested for clinical sample analyses and has been used in various studies 6,14,21,22. However, for precision of the method, investigation should be continued, focusing on the characterization based on the sequence of nucleic acids through PCR procedures (polymerase chain reaction) that point to genes of the shiga toxin (stx1 and stx2), given that BBL CHROMagar 0157 does not differentiate between the toxin producing and non-toxin producing strains of Escherichia coli 0157:H7 13.
In conclusion, the coliform bacteria had the highest presence in the samples analyzed. However, they are not considered to be the causal agent of diarrhea. The presence of Escherichia coli 0157:H7 was the second bacteria with highest percentage in the fecal samples analyzed (22.03% incidence), which does not depend on the species of the ruminant, but it represents a mortality risk factor in lactating ruminants (under 21 days of age) causing economic losses and health risk for the population of the state of Zacatecas.
REFERENCES
1. WHO. Zoonotic Non-0157 Shiga Toxin-Producing Escherichia Coli (STEC): report of a WHO scientific working group meeting, Berlin, Germany. (No. WHO/CSR/APH/98.8). Geneva: World Health Organization; 1998. https://apps.who.int/iris/bitstream/handle/10665/68880/WHO_CSR_APH_98.8.pdf;jsessionid=B1067243252676E852A98B9BF9EB08C3?sequence=1 [ Links ]
2. Méndez A, Maldonado A, Ruiz-Villamor ER, Moreno IL, Bautista MJ, Lorenzo B, et al. Enfermedades neonatales. [On line%#093; Organismo De La Unidad Nacional De Ovinocultores: México; 2016. http://www.uno.org.mx/empezar/neonatales.html [ Links ]
3. Macedo R, Arredondo V, Rodríguez J, Ramírez J, López, B. Efecto del sistema de producción, de la época de nacimiento y del sexo sobre la mortalidad neonatal de corderos Pelibuey. Trop Subtrop Agroecosyst. 2010; 12(1):77-84. http://www.revista.ccba.uady.mx/ojs/index.php/TSA/article/view/205 [ Links ]
4. Gebremedhin EZ, Agonafir A, Tessema TS, Tilahun G, Medhin G, Vitale M. et al. Some risk factors for reproductive failures and contribution of Toxoplasma gondii infection in sheep and goats of Central Ethiopia: a cross-sectional study. Res Vet Sci. 2013; 95(3):894-900. https://doi.org/10.1016/j.rvsc.2013.08.007 [ Links ]
5. Gould LH, Bopp C, Strockbine N, Atkinson R, Baselski V, Body B. et al. Recommendations for diagnosis of Shiga toxin-producing Escherichia coli infections by clinical laboratories. MMWR Recomm Rep. 2009; 58(12) 1-14. https://www.cdc.gov/mmwr/preview/mmwrhtml/rr5812a1.htm [ Links ]
6. Gutierrez ME, Janes ME, Torrico DD, Carabante KM, Prinyawiwatkul W. Assessment of the ability of five culture media for the detection of Escherichia coli O157. Int J Food Sci Technol. 2016; 51(8):1910-1915. https://doi.org/10.1111/ijfs.13164 [ Links ]
7. Laven RA, Ashmore A, Stewart CS. Escherichia coli in the rumen and colon of slaughter cattle, with particular reference to E. coli O157. Vet. J. 2003; 165 78-83. https://doi.org/10.1016/S1090-0233(02)00162-4 [ Links ]
8. Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, Harel J. et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011; 13(2):365-377. https://doi.org/10.1111/j.1462-2920.2010.02334.x [ Links ]
9. Cobbold RN. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl Environ Microbiol . 2007; 73(5):1563-1568. https://doi.org/10.1128/AEM.01742-06 [ Links ]
10. Franz E, van Hoek AH, van der Wal FJ, de Boer A, Zwartkruis-Nahuis A, van der Zwaluw K. et al. Genetic features differentiating bovine, food and human isolates of Shiga toxin-producing Escherichia coli O157 in The Netherlands. J Clin Microbiol. 2012; 50(3):772-780. http://dx.doi.org/10.1128/JCM.05964-11 [ Links ]
11. Fratamico PM, DebRoy C. Detection of Escherichia coli O157:H7 in food using real-time multiplex PCR assays targeting the stx1, stx2, wzyO157, and the fliCh7 or eae genes. Food Anal Methods. 2010; 3(4):330-337. https://doi.org/10.1007/s12161-010-9140-x [ Links ]
12. Vicente HIG, Amaral LA, Cerqueira AMF. Shigatoxigenic Escherichia coli serogroups O157, O111 and O113 in feces, water and milk samples from dairy farms. Braz J Microbiol. 2005; 36(3):217-222. http://dx.doi.org/10.1590/S1517-83822005000300003 [ Links ]
13. Hirvonen JJ, Siitonen A, Kaukoranta SS. Usability and performance of CHROMagar STEC in detection of Shiga toxin-producing Escherichia coli strains. J Clin Microbiol . 2012; 50:3586-3590. https://doi.org/10.1128/JCM.01754-12 [ Links ]
14. Etcheverria AI, Padola NL. Shiga toxin-producing Escherichia coli: factors involved in virulence and cattle colonization. Virulence. 2013; 4(5):366-372. https://doi.org/10.4161/viru.24642 [ Links ]
15. Caballero M, Rivera I, Jara LM, Ulloa-Stanojlovic FM, Shiva C. Isolation and molecular identification of potentially pathogenic Escherichia coli and Campylobacter jejuni in feral pigeons from an urban area in the city of Lima, Peru. Rev Inst Med Trop S Paulo. 2015; 57(5):393-396. http://dx.doi.org/10.1590/S0036-46652015000500004 [ Links ]
16. Heiman KE, Mody RK, Johnson SD, Griffin PM, Gould LH. Escherichia coli O157 outbreaks in the United States, 2003-2012. Emerg Infect Dis. 2015; 21(8):1293-1301. https://doi.org/10.3201/eid2108.141364 [ Links ]
17. Olvera A, Signorini M, Tarabla H. Escherichia coli verotoxigénica: modelo cuantitativo de exposición y escenarios de riesgos en canales bovinas en Argentina. Rev Panam Salud Pública. 2010; 27(6):403-413. https://doi.org/10.1590/S1020-49892010000600001 [ Links ]
18. Fox JT, Shi X, Nagaraja TG. Escherichia coli O157 in the rectoanal mucosal region of cattle. Foodborne Pathog Dis. 2008; 5(1):69-77. https://doi.org/10.1089/fpd.2008.0042 [ Links ]
19. Jacob ME, Callaway TR, Nagaraja TG. Dietary interactions and interventions affecting Escherichia coli O157 colonization and shedding in cattle. Foodborne Pathog Dis . 2009; 6(7):785-792. https://doi.org/10.1089/fpd.2009.0306 [ Links ]
20. Piedrahita D, Márquez T, Máttar S. Detección de Escherichia coli 0157: H7 en poblaciones porcinas, canal bovina y productos cárnicos en el departamento de Córdoba. Rev MVZ Córdoba. 2001; 6(2):119-126: https://doi.org/10.21897/rmvz.532 [ Links ]
21. Tang Y, Kim H, Singh AK, Aroonnual A, Bae E, Rajwa B. et al. Light scattering sensor for direct identification of colonies of Escherichia coli serogroups O26, O45, O103, O111, O121, O145 and O157. PloS One. 2014; 9(8):e105272. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0105272 [ Links ]
22. Parsons BD, Zelyas N, Berenger BM, Chui L. Detection, characterization, and typing of Shiga toxin-producing Escherichia coli. Front Microbiol. 2016; 7:478. https://doi.org/10.3389/fmicb.2016.00478 [ Links ]
How to cite (Vancouver) Lara-Duran JA, Silva-Vega M, Bañuelos-Valenzuela R, Delgadillo-Ruiz L, Delgadillo-Ruiz O. Incidence of Escherichia coli 0157:H7 in feces of lactating ruminants with diarrheal syndrome. Rev MVZ Cordoba. 2019; 24(3):7339-7345. DOI: https://doi.org/10.21897/rmvz.1232
Received: August 2018; Accepted: June 2019; Published: September 2019











 texto em
texto em