Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Revista colombiana de Gastroenterología
versão impressa ISSN 0120-9957
Rev Col Gastroenterol vol.35 no.3 Bogotá jul./set. 2020 Epub 01-Mar-2021
https://doi.org/10.22516/25007440.493
Review article
Helicobacter pylori susceptibility to six commonly used antibiotics in Colombia
1Microbiólogo y bioanalista, grupo Bacterias & Cáncer, Facultad de Medicina, Universidad de Antioquia; Medellín, Colombia
2Biólogo, grupo Bacterias & Cáncer, Departamento de Microbiología y Parasitología, Facultad de Medicina, Universidad de Antioquia; Medellín, Colombia
3Bacterióloga, grupo Bacterias & Cáncer, Facultad de Medicina, Universidad de Antioquia; Medellín, Colombia
Helicobacter pylori (H. pylori) is a microaerophilic gram-negative bacillus that colonizes the gastric mucosa. It infects more than half the world’s population, making it the most common bacterial infection. The prevalence of infection and associated diseases is high in developing countries. The recommended treatment for its eradication is triple therapy; however, its efficacy has decreased due to the lack of knowledge of the bacterial susceptibility pattern among the medical staff and the emergence of resistant strains. H. pylori susceptibility is associated with the bacteria’s ability to adapt to hostile environments and the use of antibiotics. In Colombia, it has been reported that H. pylori is resistant to amoxicillin, metronidazole, clarithromycin, furazolidone, levofloxacin, and tetracycline. Studies on the susceptibility pattern have determined that the frequency of H. pylori susceptibility is variable and demonstrate the lack of data in most of the Colombian territory. With this in mind, the objective of this review is to describe the percentage of resistance to amoxicillin, metronidazole, clarithromycin, furazolidone, levofloxacin and tetracycline, which are used for the treatment of H. pylori infection, according to studies conducted in Colombia.
Keywords: Helicobacter pylori; Antimicrobial resistance; Gastroduodenal disorders; Gastric cancer
Helicobacter pylori (H. pylori) es un bacilo gramnegativo microaerófilo, capaz de colonizar la mucosa gástrica. Este microorganismo infecta a más de la mitad de la población mundial, por lo que se ha convertido en la infección bacteriana más común. La prevalencia de la infección y de las enfermedades asociadas a ella es alta, sobre todo en países en vías de desarrollo. El tratamiento recomendado para la erradicación es la triple terapia; sin embargo, su eficacia ha disminuido por el desconocimiento del patrón de susceptibilidad bacteriano por parte del personal médico y dada la aparición de cepas resistentes. La resistencia en H. pylori se asocia con la capacidad de adaptación de la bacteria a ambientes hostiles y al uso de los antibióticos. En Colombia, existen reportes acerca de que H. pylori presenta resistencia a amoxicilina, metronidazol, claritromicina, furazolidona, levofloxacina y tetraciclina. Los estudios del patrón de susceptibilidad determinaron que la frecuencia de resistencia de H. pylori es variable y demuestran la falta de datos en la mayoría del territorio del país. Sobre la base de lo anterior, el objetivo de esta revisión es describir los porcentajes de resistencia de H. pylori a los antibióticos amoxicilina, metronidazol, claritromicina, furazolidona, levofloxacina y tetraciclina, usados en el tratamiento de la infección en los estudios realizados en Colombia.
Palabras clave: Helicobacter pylori; farmacorresistencia microbiana; enfermedades gastroduodenales; cáncer gástrico
Introduction
Helicobacter pylori (H. pylori) is a gram-negative, microaerophilic, and pleomorphic bacillus, approximately 3.5 μm in length and 0,5 μm in diameter, that affects about half of the world’s population 1,2. Infection reports vary, with a prevalence of 79.1% in Africa, 63.4% in Latin America and the Caribbean, 54.7% in Asia, 37.1% in North America, and 24.1% in Oceania 3,4.
H. pylori infection prevalence is higher in developing countries, provided that there is a well-established association between its onset, socioeconomic level and hygiene conditions 3,5. Its form of transmission is unclear, but it has been suggested that it is acquired in childhood by the fecal-oral route, and that intrafamily transmission is more frequent 6. Other studies suggest it is also transmitted by consuming contaminated water or vegetables 7,8.
In 1994, the World Health Organization (WHO), through the International Agency for Research on Cancer (IARC), classified H. pylori as a type I carcinogen 9. H. pylori Infection is the main risk factor for the development of gastric cancer (GC), with a significant positive correlation and a relative risk of 3.8 10. GC was the cause of death of 782,685 people worldwide in 2018 10,11.
Colombia has a high incidence of H. pylori infection 12. Its management is based on the international consensus of triple therapy (proton pump inhibitor, amoxicillin, and clarithromycin or metronidazole) 13. In addition, a guide for the treatment of H. pylori infections was published in 2015 in the country. However, this is not widely used by doctors and should be adjusted according to local resistance rates 14.
Therapeutic failure (TF) in H. pylori eradication is multifactorial and involves genetic, bacterial, and external factors of the patient, such as non-adherence to treatment schedule. Thus, the main cause of TF is bacterial resistance to antibiotics, which is acquired during treatment 15. Antibiotic resistance occurs because of the ability of microorganisms to adapt naturally to hostile media. It is also intrinsic to the absence of the drug binding site and acquired by genetic changes. In addition, H. pylori is capable of achieving genetic diversity by homologous mutations and recombinations 16-23. The emergence of resistant strains is also associated with the indiscriminate use of antibiotics and the lack of adherence to treatment as a result of the adverse effects derived from their use.
Currently, in Colombia, there are reports on H. pylori resistance to amoxicillin, metronidazole, clarithromycin, furazolidone, levofloxacin, moxifloxacin, and tetracycline 12,24-29. In this context, the aim of this review is to describe the patterns of resistance to these antibiotics (amoxicillin, metronidazole, clarithromycin, furazolidone, levofloxacin and tetracycline) that have been described in the treatment of H. pylori infection by studies conducted in Colombia.
Materials and methods
Search Strategy and Selection Criteria
Searches were performed in the PubMed (National Library of Medicine of the United States, Bethesda, MD), LILACS (Latin American Literature of Information in Health Sciences: http://lilacs.bvsalud.org/en) and SciELO (electronic scientific library: http://www.scielo.org) databases. Articles were searched using the following strategy: Helicobacter pylori, AND Resistencia AND Colombia in Spanish, and Helicobacter pylori, AND drug resistance AND Colombia in English.
Inclusion Criteria
Original full-text available articles describing on H. pylori resistance to antibiotics in Colombia, regardless of their year of publication, were selected. From these studies, information such as the first author, the year of publication, the place where the research was conducted, the year samples were collected, the antibiotics evaluated, the number of samples (gastric biopsies or H. pylori isolates), the prevalence of antibiotic resistance, and the resistance assessment method (epsilometry test [E-test], disk diffusion or polymerase chain reaction mutation detection [Polymerase Chain Reaction, PCR]) were obtained.
Results
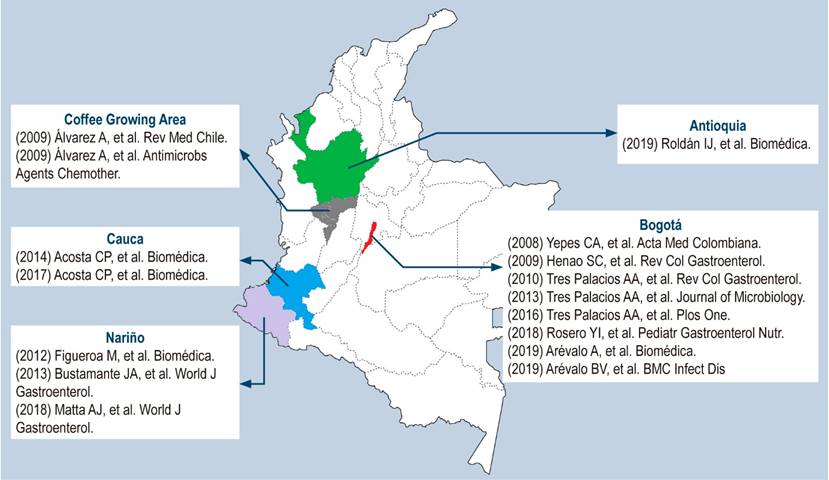
The search strategy in Spanish and English yielded 39 articles. Of these, 23 met the inclusion criteria. The following is a flowchart of the search and the selection process of the studies finally included in the review after the search strategy was applied (Figure 1). In addition, a summary table with the studies on H. pylori resistance conducted in Colombia is also shown (Table 1).
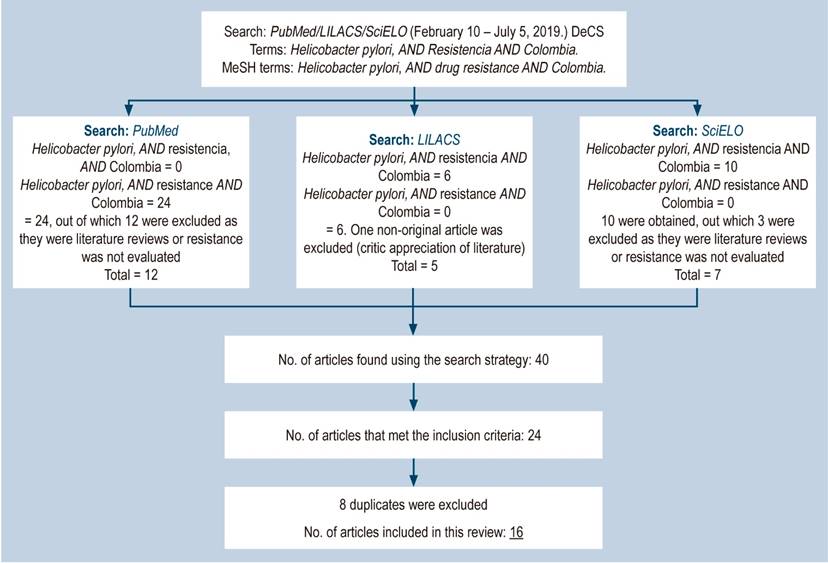
Figure 1 General outline of the review strategy on Helicobacter pylori antimicrobial resistance in Colombia. DeCS: Health Science Descriptors; MeSH: Medical Subject Headings.
Table 1 Summary Table with H. pylori resistance studies conducted in Colombia
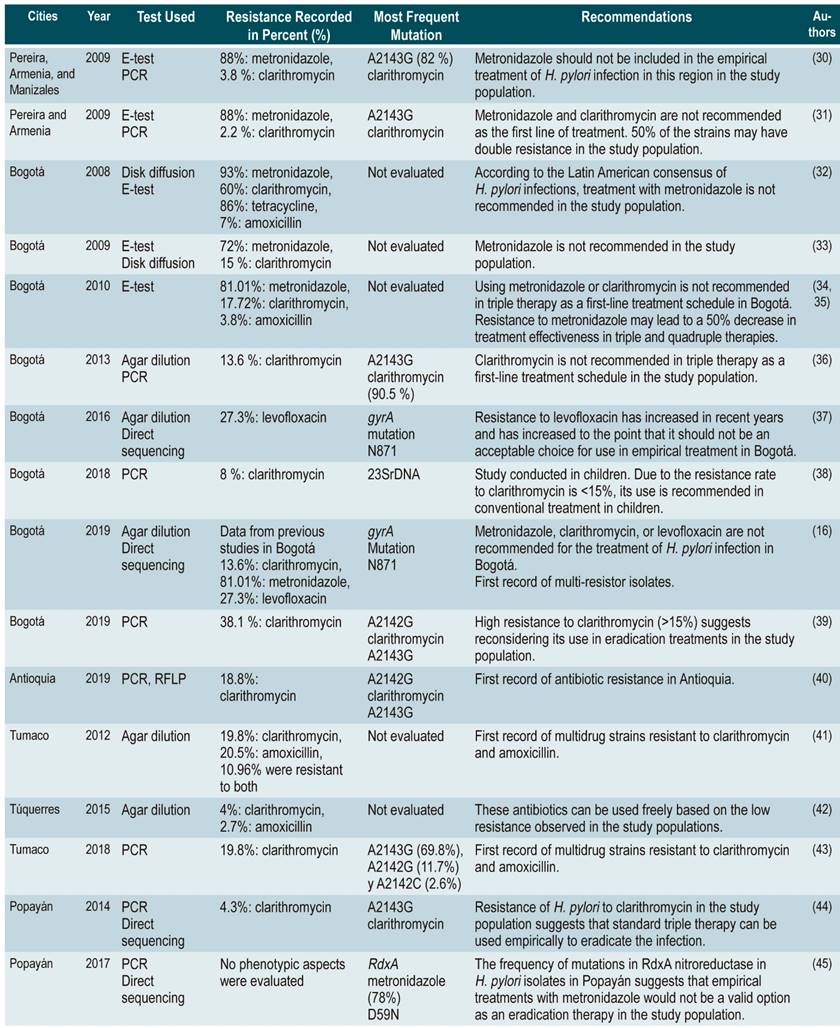
E-test: Epsilometry test; PCR: Polymerase chain reaction; RFLP: Restriction Fragment Length Polymorphisms.
Amoxicillin
Amoxicillin is a first-choice antibiotic to treat H. pylori infection due to the high sensitivity of the microorganism. Amoxicillin is a semi-synthetic penicillin that belongs to beta-lactams and is an inhibitor of peptidoglycan synthesis. It blocks penicillin-binding protein (PBP) transporters 13,30. H. pylori becomes resistant to amoxicillin through mutations in the pbpa gene (PBP), the production of β-lactamases, and the presence of efflux pumps. The last two factors are associated with the highest resistance to this drug 25,31.
The most frequent mutations in the pbpa gene substitutions are observed in specific positions: a) in amino acid 556, substitution from serine to threonine (Thr556→SER) takes place; b) in position 648, a substitution from lysine to a glycine (Lys648→Gly) occurs; c) in position 649, an arginine to lysine (Arg649→Lys) substitution, and d) in position 656, an arginine to proline (Arg656→Pro) substitution.
The first strains of H. pylori resistant to amoxicillin were recorded in 1997 in a patient who had previously been treated 32. In 2009, the appearance of a β-lactamases producing strain (TEM-1) capable of destroying the beta-lactamic ring of the antibiotic was described 25. In addition, efflux pumps were reported on the bacterial wall, expelling the antibiotic to the outside of the cell. Although this was only observed in in-vitro strains, it is of great concern as the pumps cause the strains to become multi-resistant 31.
Camargo et al., in a review conducted in 2014 and in which several studies on H. pylori resistance conducted in Latin America were included, found that Colombia and Brazil had the highest rates of amoxicillin resistance: 7 and 15%, respectively 12. In addition, in 2007, Gómez et al., in a study were 648 physicians (68%: General physicians, 19%: Internist physicians, and 13%: Gastroenterologists) from 9 Colombian cities were surveyed, reported that amoxicillin was the most prescribed antibiotic to treat H. pylori infection, with a 73% rate 33. In Colombia, studies show H. pylori resistance rates to amoxicillin of 1.9%, 3.8%, and 9.5% 34-36.
Metronidazole
The bactericidal action of metronidazole depends on enzyme reduction. In the case of H. pylori, which does not have the superoxide dismutase enzyme, the decrease effect of metronidazole is achieved through NADPH (nicotinamide adenine dinucleotide phosphate) nitroreductase and NAD(P)H flavin oxidoreductase to produce anionic radicals as nitrous derivatives and hydroxylamines, which lead to inhibition in the synthesis of nucleic acids when bound to the imidazole ring 37-39. The resistance mechanisms acquired by the bacteria point mainly to mutations in the RdxA gene, which encodes NADPH nitroreductase, and FdxB, which encodes ferredoxin 38,39.
Some authors state that point mutations in RdxA and, less frequently, changes in FrxA. cause resistance to metronidazole Others claim that FrxA mutations only boost resistance caused by the inactivation of RdxA. Although mutations occur with RdxA function loss, not always such function loss implies a decrease in susceptibility to the drug, which leads to the fact that genotypic results obtained are not consistent with those in-vitro and in-vivo results40.
In addition, the agar dilution method and the epsilometer test (E-test) present inter- and intra-test variability and are not accurate, probably because of the environmental conditions necessary for the culture of H. pylori 39,41. Regarding the epsilometer test, some authors argue that it overestimates the resistance to metronidazole, and therefore propose to support the resistance diagnosis with molecular biology tests.
In Latin America, the reported resistance to metronidazole is >30%. However, in countries such as Peru and Colombia, the average documented resistance is 66 and 83%, respectively 12. In Colombia, different studies describe resistance rates to metronidazole, through microbiological methods, ranging from 72 to 97.6% 34-36. Also, in 2017, Acosta et al. evaluated the frequency of RdxA mutations by genotypic methods and found that 78.2% of cases (133/170) showed some genetic alteration associated with drug resistance 42. Metronidazole is one of the most prescribed antimicrobials by general practitioners in Colombia to treat H. pylori infection; thus, high resistance rates seem to be ignored 33.
Clarithromycin
Clarithromycin is a macrolide with bacteriostatic and bactericidal activity that inhibits protein synthesis by binding to the 23S component of the 50S ribosomal subunit 43. H. pylori becomes resistant to clarithromycin through mutations in the peptide-diltransferase region of the V domain of the HPrrnB23S gene, which encodes the 23S ribosomal RNA 43. The most frequent mutations in this gene are located at positions 2142, with the transition of an adenine to a guanine (A2142G) and the adenine-cytosine transversion (A2142C.) The adenine-to-guanine transition at position 2143 (A2143G) produces a conformal change in the antibiotic binding site and prevents its action 44,45. Other less frequent mutations exist, including T2182C 46, T2717C 47, G2224A, C2245T, and T2289C transitions 48.
In Latin America, a prevalence of macrolide resistance of 14% and 13% has been reported in Argentina and Mexico, respectively 12. In Colombia, a study conducted in 2009 in the Coffee Growing Region showed low resistance to clarithromycin (2.2%). However, this antibiotic is used only to treat respiratory diseases in childhood, which would explain the difference between the percentages found 33,35.
On the other hand, studies on the prevalence of macrolide resistance in Colombia describe values ranging from 13.6 to 63.1% 29,34,36,49. As for genotypic resistance, it has been determined that the transition of adenine to guanine at position 2143 (A2143G) is the most frequent (90.5%), followed by the A2142G (7.1%) and A2142C (2.4%) transitions 29.
Furazolidone
Furazolidone is one of the nitrofurans proposed to treat H. pylori infection as salvage therapy. Nitrofurans are bacteriostatic, but at high doses they are bactericidal, and act similarly to nitroimidazoles. Little is known regarding the resistance mechanism to this group of antibiotics. It has been suggested that pyruvate-flavodoxin oxidoreductase (encoded by the gene PorCDAB) and 2-oxoglutarate oxidoreductase (OorDABC) are the nitroreductases involved in this process 50.
Su et al. identified mutations associated with furazolidone resistance in the porD gene, with guanine-adenine transitions at position 353 (G353A), adenine-guanine at position 356 (A356G), and cytokine-thymine at position 357 (C357T), and in oorD, with adenine transitions (A041G, A122G) and transversion (C349A.) However, some drug-resistant strains did not carry mutations, so there is a possibility of other mutations that have not yet been determined 51. Evidence suggests that other enzymes would activate nitrofurans; however, there are few studies on this subject 52.
In Colombia, there is only one phenotypic resistance study on H. pylori, which in fact is a thesis, describing a resistance of 4.8%. Beyond that, there are no other studies on genotypic resistance to nitrofurans conducted in the country 33,53 Therefore, the prescription of nitrofurans is very low and are recommended as a rescue therapy in countries such as Brazil, where a resistance of 3% has been reported, a similar figure to that found in Bogotá 12.
Levofloxacin
Levofloxacin is a fluoroquinolone used in some cases as rescue therapy in unsuccessful treatments with clarithromycin. Fluoroquinolones act by binding to subunit A of DNA gyrase and prevent the formation of tetramer (two subunits A and two B), thereby blocking the function of this enzyme. Subunits A and B are coded by genes gyrA and gyrB, respectively 37. The resistance of H. pylori to fluoroquinolones is caused by mutations in gyrA and gyrB, and by alterations in porins.
In this way, mutations occur in the Quinolone Resistance Domain Region (QRDR) region of gyrA, which prevents antibiotic binding to gyrase, by altering the quinolone binding site in the DNA-gyrase DNA complex 54. Mutations described as causing this phenomenon are unique mutations: Asn87 → Lys, Ala88 → Val, Asp91 → Gly, Tyr or Asn, and double mutations in Asp91 → Asn and in Ala88 → Val 55.
In Colombia, a 2009-2014 follow-up study conducted in Bogotá that evaluated levofloxacin resistance in biopsies from patients who underwent endoscopy, found that resistance in 2009 was 11.8%, while in 2014 it was 27.3%. These authors considered that this increase is due to the use of this drug for treating respiratory and urinary tract infections 56.
Tetracyclines
Tetracyclines are used in quadruple therapy regimens for the eradication of H. pylori13. These have a bacteriostatic effect when they bind reversibly to the 30S ribosomal subunit and inhibit protein synthesis. The mutations described are found in the HPrrnA-16S gene, which codes for the 16S component of the minor subunit (30S). The most common involve nucleotides 926-928 (AGA→TTC). A926T/A928C, A926G/G927T, A926G/A928C double mutations and point mutation at the A926G transition or the A939C transversion have also been described 57-61.
In studies conducted in Latin America, low tetracycline resistance rates have been reported, ranging from 6 to 14% 12. However, for Colombia, Yepes et al. (2018) reported that 85.7% (72/84) of the strains isolated in their study showed resistance to this drug 34. This data differs greatly from records in the rest of the continent.
Discussion
According to the percentages of H. pylori resistance in Colombia, metronidazole stands out as the antibiotic that causes the greatest resistance in the bacteria (Table 1). In Bogotá, the recorded resistance values ranged from 72 to 93% 34,36,62, while in the Coffee Growing Region, it was 88% 35. The only genotypic study was conducted in Popayán, where the most frequent mutation was found in D59N (78%) 42. Different authors recommend avoiding using metronidazole to treat H. pylori infection in the cities where their studies were conducted. Due to the lack of more information, it is not possible to determine the situation in the rest of the country.
Clarithromycin is the antibiotic with the highest number of resistance studies conducted in Colombia, showing significant differences. In the case of Bogotá, resistance rates to this drug have varied considerably between studies: 13.6% and 60% 29,34,36,62. In Tumaco, the resistance rate was 19.8% 63, while in Armenia and Pereira it was lower, 2.2% 35 and 3.8%, respectively 64.
In Popayán, the genotypic resistance to clarithromycin prevalence was 4.3%, and the A2143G transition was the most frequent 65, whereas in Antioquia it was 18.8%, where the frequency of the A2143G transition was 81.5% 49.
Similarly, resistance to amoxicillin was assessed in 3 studies, 2 of which were conducted in Bogotá. Resistance values were 7% in 2008 34 and 3.8% in 2010 36. The third study was carried out in Tumaco and found a resistance of 20.5% 63.
With regard to levofloxacin resistance values in Colombia, 11.8% of the strains studied in Bogotá during 2009 and 27.3% in 2014 were resistant to this drug 56. The authors of said study suggest that levofloxacin should not be considered as an antibiotic of choice for empirical therapy of H. pylori infection. Likewise, resistance to tetracycline was 85.7%, which is reported by the only study that has been developed in our country on the resistance of H. pylori to this drug 34.
On the other hand, two studies have shown the emergence of multidrug-resistant H. pylori strains. The first was carried out in Tumaco in 2012 and detected the presence of strains resistant to clarithromycin and amoxicillin 63, while the second, conducted by Arévalo et al. in 2019, described strains resistant to 2 or more antibiotics (amoxicillin, clarithromycin, levofloxacin and metronidazole) These strains were found in isolates obtained from patients with 3 or 4 failed treatments 16.
According to the clinical practice guidelines for the diagnosis and treatment of H. pylori infection in adults, in Colombia, avoiding the use triple combination therapy of amoxicillin, clarithromycin, and metronidazole or levofloxacin as first-line therapy is recommended, provided that evidence shows the primary resistance of H. pylori to this type of therapy. Therefore, local epidemiology should be the basis to suggest an adjusted and appropriate treatment and avoid using antibiotics empirically that may lead to therapeutic failure 14.
The data presented in this review show there are variations between populations, as well as the importance of determining the resistance pattern of each population in the country. This must be done in order to monitor the frequency in which the different resistances of the strains present in the country occur.
It is worth noting that the data obtained from the studies apply only to the study population and should not be extrapolated to the country as a whole. The following map is a graphical representation of the studies that were included in this review (Figure 2).
Thus, the findings of this review show the need for further studies to determine H. pylori resistance to antibiotics in the Colombian population in order to define the antibiotics that must be prescribed as first-line or rescue therapy in the country.
REFERENCES
1. Loughlin MF. Novel therapeutic targets in Helicobacter pylori. Expert Opin Ther Targets. 2003;7(6):725-735. http://doi.org/10.1517/14728222.7.6.725 [ Links ]
2. Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449-490. http://doi.org/10.1128/CMR.00054-05 [ Links ]
3. O’Connor A, O’Morain CA, Ford AC. Population screening and treatment of Helicobacter pylori infection. Nat Rev Gastroenterol Hepatol. 2017;14(4):230-240. http://doi.org/10.1038/nrgastro.2016.195 [ Links ]
4. Silva GM, Silva HM, Nascimento J, Gonçalves JP, Pereira F, Lima R. Helicobacter pylori antimicrobial resistance in a pediatric population. Helicobacter. 2018;23(5):e12528. http://doi.org/10.1111/hel.12528 [ Links ]
5. Nagy P, Johansson S, Molloy-Bland M. Systematic review of time trends in the prevalence of Helicobacter pylori infection in China and the USA. Gut Pathog. 2016;8:8. http://doi.org/10.1186/s13099-016-0091-7 [ Links ]
6. Goh KL, Chan WK, Shiota S, Yamaoka Y. Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter. 2011;16 Suppl 1(0 1):1-9. http://doi.org/10.1111/j.1523-5378.2011.00874.x [ Links ]
7. Fernández-Delgado M, Contreras M, García-Amado MA, Michelangeli F, Suárez P. Evidencias de la transmisión acuática de Helicobacter pylori. Interciencia. 2008;33(6):412-417. [ Links ]
8. Gomes BC, de Martinis EC. Fate of Helicobacter pylori artificially inoculated in lettuce and carrot samples. Braz J Microbiol. 2004;35(1):145-150. https://doi.org/10.1590/S1517-83822004000100024 [ Links ]
9. International Agency for Research on Cancer (IARC). Helicobacter pylori eradication as a strategy for preventing gastric cancer: IARC working group report, volume 8. Lyon: International Agency for Research on Cancer; 2014. [ Links ]
10. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. http://doi.org/10.3322/caac.21492 [ Links ]
11. IARC Working Group on the Evaluation of Carcinogenic Risk to Humans. Schistosomes, Liver Flukes and Helicobacter pylori. Lyon (FR): International Agency for Research on Cancer; 1994. (IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, No. 61). Disponible en: https://www.ncbi.nlm.nih.gov/books/NBK487782/ [ Links ]
12. Camargo MC, García A, Riquelme A, Otero W, Camargo CA, Hernández-García T, Candia R, Bruce MG, Rabkin CS. The problem of Helicobacter pylori resistance to antibiotics: a systematic review in Latin America. Am J Gastroenterol. 2014;109(4):485-95. http://doi.org/10.1038/ajg.2014.24 [ Links ]
13. Malfertheiner P, Megraud F, O’Morain CA, Gisbert JP, Kuipers EJ, Axon AT, Bazzoli F, Gasbarrini A, Atherton J, Graham DY, Hunt R, Moayyedi P, Rokkas T, Rugge M, Selgrad M, Suerbaum S, Sugano K, El-Omar EM; European Helicobacter and Microbiota Study Group and Consensus panel. Management of Helicobacter pylori infection-the Maastricht V/Florence Consensus Report. Gut. 2017;66(1):6-30. http://doi.org/10.1136/gutjnl-2016-312288 [ Links ]
14. Otero W, Trespalacios AA, Otero L, Vallejo MT, Torres M, Pardo R, Sabbagh L. Guía de práctica clínica para el diagnóstico y tratamiento de la infección por Helicobacter pylori en adultos. Rev Col Gastroenterol. 2015;30 supl. 1:17-33. [ Links ]
15. Wang D, Guo Q, Yuan Y, Gong Y. The antibiotic resistance of Helicobacter pylori to five antibiotics and influencing factors in an area of China with a high risk of gastric cancer. BMC Microbiol. 2019;19(1):152. http://doi.org/10.1186/s12866-019-1517-4 [ Links ]
16. Arévalo A, Otero WA, Trespalacios AA. Helicobacter pylori: resistencia múltiple en pacientes de Bogotá, Colombia. Biomédica. 2019;39 supl. 1:125-134. https://doi.org/10.7705/biomedica.v39i3.4437 [ Links ]
17. Björkholm B, Sjölund M, Falk PG, Berg OG, Engstrand L, Andersson DI. Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc Natl Acad Sci U S A. 2001;98(25):14607-14612. http://doi.org/10.1073/pnas.241517298 [ Links ]
18. Jorgensen M, Daskalopoulos G, Warburton V, Mitchell HM, Hazell SL. Multiple strain colonization and metronidazole resistance in Helicobacter pylori-infected patients: identification from sequential and multiple biopsy specimens. J Infect Dis. 1996;174(3):631-635. http://doi.org/10.1093/infdis/174.3.631 [ Links ]
19. Enroth H, Björkholm B, Engstrand L. Occurence of resistance mutation and clonal expansion in Helicobacter pylori multiple-strain infection: a potential risk in clarithromycin-based therapy. Clin Infect Dis. 1999;28(6):1305-1307. http://doi.org/10.1086/514796 [ Links ]
20. Otero W, Trespalacios AA, Otero E. Helicobacter pylori: Tratamiento actual, un importante reto en gastroenterología. Rev Col Gastroenterol. 2009;24(3):279-292. [ Links ]
21. Furuta Y, Konno M, Osaki T, Yonezawa H, Ishige T, Imai M, Shiwa Y, Shibata-Hatta M, Kanesaki Y, Yoshikawa H, Kamiya S, Kobayashi I. Microevolution of Virulence-Related Genes in Helicobacter pylori Familial Infection. PLoS One. 2015;10(5):e0127197. http://doi.org/10.1371/journal.pone.0127197 [ Links ]
22. Cao Q, Didelot X, Wu Z, Li Z, He L, Li Y, Ni M, You Y, Lin X, Li Z, Gong Y, Zheng M, Zhang M, Liu J, Wang W, Bo X, Falush D, Wang S, Zhang J. Progressive genomic convergence of two Helicobacter pylori strains during mixed infection of a patient with chronic gastritis. Gut. 2015;64(4):554-61. http://doi.org/10.1136/gutjnl-2014-307345 [ Links ]
23. Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, Dyrek I, Achtman M. Free recombination within Helicobacter pylori. Proc Natl Acad Sci U S A. 1998;95(21):12619-24. http://doi.org/10.1073/pnas.95.21.12619 [ Links ]
24. Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance?. Nat Rev Microbiol. 2010;8(4):260-271. http://doi.org/10.1038/nrmicro2319 [ Links ]
25. Tseng YS, Wu DC, Chang CY, Kuo CH, Yang YC, Jan CM, Su YC, Kuo FC, Chang LL. Amoxicillin resistance with beta-lactamase production in Helicobacter pylori. Eur J Clin Invest. 2009;39(9):807-12. http://doi.org/10.1111/j.1365-2362.2009.02166.x [ Links ]
26. Binh TT, Suzuki R, Trang TT, Kwon DH, Yamaoka Y. Search for novel candidate mutations for metronidazole resistance in Helicobacter pylori using next-generation sequencing. Antimicrob Agents Chemother. 2015;59(4):2343-2348. http://doi.org/10.1128/AAC.04852-14 [ Links ]
27. Wang LH, Cheng H, Hu FL, Li J. Distribution of gyrA mutations in fluoroquinolone-resistant Helicobacter pylori strains. World J Gastroenterol. 2010;16(18):2272-2277. http://doi.org/10.3748/wjg.v16.i18.2272 [ Links ]
28. Talebi Bezmin Abadi A, Ghasemzadeh A, Taghvaei T, Mobarez AM. Primary resistance of Helicobacter pylori to levofloxacin and moxifloxacine in Iran. Intern Emerg Med. 2012;7(5):447-452. http://doi.org/10.1007/s11739-011-0563-1 [ Links ]
29. Trespalacios AA, Otero W, Caminos JE, Mercado MM, Avila J, Rosero LE, Arévalo A, Poutou-Piñales RA, Graham DY. Phenotypic and genotypic analysis of clarithromycin-resistant Helicobacter pylori from Bogotá D.C., Colombia. J Microbiol. 2013;51(4):448-52. http://doi.org/10.1007/s12275-013-2465-6 [ Links ]
30. Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20(2):280-322. http://doi.org/10.1128/CMR.00033-06 [ Links ]
31. Liu ZQ, Zheng PY, Yang PC. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J Gastroenterol. 2008;14(33):5217-5222. http://doi.org/10.3748/wjg.14.5217 [ Links ]
32. Dore MP, Sepúlveda AR, Mura I, Realdi G, Osato MS, Graham DY. Explanation for variability of omeprazole amoxycillin therapy? Tolerance of H. pylori to amoxycillin. Gastroenterology.1997;112:A105. [ Links ]
33. Gómez M, Otero W, Gutiérrez O. Tratamiento de la infección por Helicobacter pylori. Encuesta en un grupo de médicos generales y especialistas en Colombia. Rev Col Gastroenterol. 2007;22(1):7-16. [ Links ]
34. Yepes CA, Rodríguez A, Ruiz A, Ariza B. Resistencia antibiótica del Helicobacter pylori en el Hospital Universitario San Ignacio de Bogotá. Acta Med Colomb. 2008;33(1):11-14. [ Links ]
35. Álvarez A, Moncayo JI, Santacruz JJ, Corredor LF, Reinosa E, Martínez JW, Beltrán L. Resistencia a metronidazol y claritromicina en aislamientos de Helicobacter pylori de pacientes dispépticos en Colombia. Red Med Chile. 2009;137(10):1309-1314. http://dx.doi.org/10.4067/S0034-98872009001000005 [ Links ]
36. Trespalacios AA, Otero W, Mercado M. Resistencia de Helicobacter pylori a metronidazol, claritromicina y amoxicilina en pacientes colombianos. Rev Col Gastroenterol. 2010;25(1):31-38. [ Links ]
37. Francesco VD, Zullo A, Hassan C, Giorgio F, Rosania R, Ierardi E. Mechanisms of Helicobacter pylori antibiotic resistance: An updated appraisal. World J Gastrointest Pathophysiol. 2011;2(3):35-41. http://doi.org/10.4291/wjgp.v2.i3.35 [ Links ]
38. Connor A, Vaira D, Gisbert JP, O’Morain C. Treatment of Helicobacter pylori Infection 2014. Helicobacter. 2014;19(1):38-45. https://doi.org/10.1111/hel.12163 [ Links ]
39. Jenks PJ, Edwards DI. Metronidazole resistance in Helicobacter pylori. Int J Antimicrob Agents. 2002;19(1):1-7. http://doi.org/10.1016/s0924-8579(01)00468-x [ Links ]
40. Kim SY, Joo YM, Lee HS, Chung IS, Yoo YJ, Merrell DS, Cha JH. Genetic analysis of Helicobacter pylori clinical isolates suggests resistance to metronidazole can occur without the loss of functional rdxA. J Antibiot (Tokyo). 2009;62(1):43-50. http://doi.org/10.1038/ja.2008.6 [ Links ]
41. Osato MS, Reddy R, Reddy SG, Penland RL, Graham DY. Comparison of the Etest and the NCCLS-approved agar dilution method to detect metronidazole and clarithromycin resistant Helicobacter pylori. Int J Antimicrob Agents. 2001;17(1):39-44. http://doi.org/10.1016/s0924-8579(00)00320-4 [ Links ]
42. Acosta CP, Quiroga AJ, Sierra CH, Trespalacios AA. Frecuencia de mutaciones de la nitrorreductasa RdxA de Helicobacter pylori para la activación del metronidazol en una población del departamento del Cauca, Colombia. Biomédica. 2017;37(2):191-199. https://doi.org/10.7705/biomedica.v37i2.3007 [ Links ]
43. Versalovic J, Shortridge D, Kibler K, Griffy MV, Beyer J, Flamm RK, Tanaka SK, Graham DY, Go MF. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40(2):477-80. http://doi.org/10.1128/AAC.40.2.477 [ Links ]
44. Mégraud F, Corti R. Resistencia bacteriana del Helicobacter pylori en el mundo en el año 2009. Acta Gastroenterol Latinoam. 2009;39(4):282-290. [ Links ]
45. Schmitt BH, Regner M, Mangold KA, Thomson RB Jr, Kaul KL. PCR detection of clarithromycin-susceptible and -resistant Helicobacter pylori from formalin-fixed, paraffin-embedded gastric biopsies. Mod Pathol. 2013;26(9):1222-1227. http://doi.org/10.1038/modpathol.2013.48 [ Links ]
46. Agudo S, Pérez-Pérez G, Alarcón T, López-Brea M. High prevalence of clarithromycin-resistant Helicobacter pylori strains and risk factors associated with resistance in Madrid, Spain. J Clin Microbiol. 2010;48(10):3703-3707. http://doi.org/10.1128/JCM.00144-10 [ Links ]
47. Fontana C, Favaro M, Minelli S, Criscuolo AA, Pietroiusti A, Galante A, Favalli C. New site of modification of 23S rRNA associated with clarithromycin resistance of Helicobacter pylori clinical isolates. Antimicrob Agents Chemother. 2002;46(12):3765-9. http://doi.org/10.1128/aac.46.12.3765-3769.2002 [ Links ]
48. Hao Q, Li Y, Zhang ZJ, Liu Y, Gao H. New mutation points in 23S rRNA gene associated with Helicobacter pylori resistance to clarithromycin in northeast China. World J Gastroenterol. 2004;10(7):1075-1077. http://doi.org/10.3748/wjg.v10.i7.1075 [ Links ]
49. Roldán IJ, Castaño R, Navas MC. Mutaciones del gen ARN ribosómico 23S de Helicobacter pylori asociadas con resistencia a claritromicina en pacientes atendidos en una unidad de endoscopia de Medellín, Colombia. Biomédica. 2019;39(supl. 2):117-129. https://doi.org/10.7705/biomedica.v39i4.4377 [ Links ]
50. Kwon DH, Lee M, Kim JJ, Kim JG, El-Zaatari FA, Osato MS, Graham DY. Furazolidone- and nitrofurantoin-resistant Helicobacter pylori: prevalence and role of genes involved in metronidazole resistance. Antimicrob Agents Chemother. 2001;45(1):306-8. http://doi.org/10.1128/AAC.45.1.306-308.2001 [ Links ]
51. Su Z, Xu H, Zhang C, Shao S, Li L, Wang H, Wang H, Qiu G. Mutations in Helicobacter pylori porD and oorD genes may contribute to furazolidone resistance. Croat Med J. 2006;47(3):410-5. [ Links ]
52. Sisson G, Goodwin A, Raudonikiene A, Hughes NJ, Mukhopadhyay AK, Berg DE, Hoffman PS. Enzymes associated with reductive activation and action of nitazoxanide, nitrofurans, and metronidazole in Helicobacter pylori. Antimicrob Agents Chemother. 2002;46(7):2116-23. http://doi.org/10.1128/aac.46.7.2116-2123.2002 [ Links ]
53. Gambia CR, Quevedo NA. Determinación in vitro de la resistencia de Helicobacter pylori a la furazolidona. Bogotá: Facultad de Ciencias, Pontificia Universidad Javeriana; 2009. [ Links ]
54. Glocker E, Kist M. Rapid detection of point mutations in the gyrA gene of Helicobacter pylori conferring resistance to ciprofloxacin by a fluorescence resonance energy transfer-based real-time PCR approach. J Clin Microbiol. 2004;42(5):2241-2246. http://doi.org/10.1128/jcm.42.5.2241-2246.2004 [ Links ]
55. Tankovic J, Lascols C, Sculo Q, Petit JC, Soussy CJ. Single and double mutations in gyrA but not in gyrB are associated with low- and high-level fluoroquinolone resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2003;47(12):3942-3944. http://doi.org/10.1128/aac.47.12.3942-3944.2003 [ Links ]
56. Trespalacios-Rangel AA, Otero W, Arévalo-Galvis A, Poutou-Piñales RA, Rimbara E, Graham DY. Surveillance of Levofloxacin Resistance in Helicobacter pylori Isolates in Bogotá-Colombia (2009-2014). PLoS One. 2016;11(7):e0160007. http://doi.org/10.1371/journal.pone.0160007 [ Links ]
57. Ribeiro ML, Gerrits MM, Benvengo YH, Berning M, Godoy AP, Kuipers EJ, Mendonça S, van Vliet AH, Pedrazzoli J Jr, Kusters JG. Detection of high-level tetracycline resistance in clinical isolates of Helicobacter pylori using PCR-RFLP. FEMS Immunol Med Microbiol. 2004;40(1):57-61. http://doi.org/10.1016/S0928-8244(03)00277-3 [ Links ]
58. Gerrits MM, Berning M, Van Vliet AH, Kuipers EJ, Kusters JG. Effects of 16S rRNA gene mutations on tetracycline resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2003;47(9):2984-2986. http://doi.org/10.1128/aac.47.9.2984-2986.2003 [ Links ]
59. Wu JY, Kim JJ, Reddy R, Wang WM, Graham DY, Kwon DH. Tetracycline-resistant clinical Helicobacter pylori isolates with and without mutations in 16S rRNA-encoding genes. Antimicrob Agents Chemother. 2005;49(2):578-583. http://doi.org/10.1128/AAC.49.2.578-583.2005 [ Links ]
60. Toledo H, López-Solís R. Tetracycline resistance in Chilean clinical isolates of Helicobacter pylori. J Antimicrob Chemother 2010;65(3):470-473. https://doi.org/10.1093/jac/dkp457 [ Links ]
61. Lawson AJ, Elviss NC, Owen RJ. Real-time PCR detection and frequency of 16S rDNA mutations associated with resistance and reduced susceptibility to tetracycline in Helicobacter pylori from England and Wales. J Antimicrob Chemother. 2005;56(2):282-286. http://doi.org/10.1093/jac/dki199 [ Links ]
62. Henao SC, Otero W, Ángel LA, Martínez JD. Resistencia primaria a metronidazol en aislamientos de Helicobacter pylori en pacientes adultos de Bogotá, Colombia. Rev Col Gastroenterol. 2009;24(1):10-15. [ Links ]
63. Figueroa M, Cortés A, Pazos A, Bravo LE. Sensibilidad in vitro a amoxicilina y claritromicina de Helicobacter pylori obtenido de biopsias gástricas de pacientes en zona de bajo riesgo para cáncer gástrico. Biomédica. 2012;32(1):32-42. https://doi.org/10.7705/biomedica.v32i1.454 [ Links ]
64. Álvarez A, Moncayo JI, Santacruz JJ, Santacoloma M, Corredor LF, Reinosa E. Antimicrobial susceptibility and mutations involved in clarithromycin resistance in Helicobacter pylori isolates from patients in the western central region of Colombia. Antimicrob Agents Chemother. 2009;53(9):4022-4024. http://doi.org/10.1128/AAC.00145-09 [ Links ]
65. Acosta CP, Hurtado FA, Trespalacios AA. Determinación de mutaciones de un solo nucleótido en el gen 23S rRNA de Helicobacter pylori relacionadas con resistencia a claritromicina en una población del departamento del Cauca, Colombia. Biomédica. 2014;34(Supl. 1):156-62. http://dx.doi.org/10.7705/biomedica.v34i0.1649 [ Links ]
Citation: Atehortúa-Rendón JD, Martínez A, Pérez-Cala TL. Helicobacter pylori susceptibility to six commonly used antibiotics in Colombia. Rev Colomb Gastroenterol. 2020;35(3):351-361. https://doi.org/10.22516/25007440.493
Received: December 24, 2019; Accepted: June 11, 2020











 texto em
texto em