1 Introduction
Antibiotics are widely used in the livestock industry for treatment of infections, disease prevention (prophylaxis), and as growth promoters in subtherapeutic dosage. Their use began during the fifties in response to the rapidly growing demand of meat worldwide [1]. Nevertheless, it is now well known that antibiotic use in livestock selects resistant pathogens, which may become an overlapped resistance reservoir between animals and humans, ensuing spread in the environment and causing an impact in public health [2].
The first evidence of this reality emerged in 1994, when glycopeptide resistant enterococcus (GRE) were isolated from livestock in Great Britain, Germany and Denmark [3],[4],[5],[2]. Thereafter, the occurrence of GRE infections increased dramatically in hospitals all around Europe and the United States, followed by the fact that these bacteria did not respond to any antibiotics available then, making GRE infections virtually untreatable.
The European Union banned the use of some antibiotics in livestock production, creating a unique opportunity for scientists to research into the effects of selective pressure removal on antimicrobial resistance spread and occurrence [2]. Results proved that after antimicrobial use in livestock ceased, the prevalence of antibiotic-resistant pathogens was significantly reduced, with little to no changes in productivity and animal health [6],[2]. In this context, there has been a fast emergence of alternatives to antibiotics in livestock production within the last three decades [1]. One promising alternative consists in the use of probiotics as prevention tools from infectious diseases, and as growth promoters.
Probiotics are live microorganisms, for the most part bacteria that once ingested, bring about health benefits beyond their inherent nutritional value [7]. These microorganisms have physiological characteristics that allow them to modify the ecology and the functional properties of the gut microbiome, which in turn, plays a fundamental role in preserving the homeostasis of the digestive system [8].
The gut of domestic animals hosts a wide variety of dense and complex microbial communities composed of bacteria, yeast, protozoa, archaea, and viruses [8]. The range of activities performed by the gut microbiota (GM) is also wide and diverse: vitamin synthesis, bioconversion of toxic compounds, immune system stimulation, and maintenance of gut peristalsis. It also plays a role as a barrier that blocks pathogen colonization of the intestine walls [8],[9].
In the last four decades, a considerable amount of research has been committed to the characterization of the digestive ecosystem. These efforts have led to the description of key bacteria that play a role in the healthy functioning of GM [8],[9].
The possibility of using key bacteria in animal feed as probiotics, that would potentially help to maintain a pathogen resistant gut, has produced considerable interest during the last few years; also taking into account the spread of antibiotic-resistant pathogens and the presence of chemical growth promoters left in meat from substances added to animal feed [8].
The most commonly used probiotics in monogastric animal (birds and pigs) feed are bacteria belonging to the genus Lactobacillus, Enterococcus, Pediococcus, and Bacillus [8]. Multiple effects have been attributed to their use in poultry. In broilers, an increased resistance to infectious diseases produced by Salmonella, Escherichia coli, and Clostridium perfringens has been described [10],[11],[12],[13],[14],[15], as well as an increased bone health and fat percentage [16], and a reduction of behaviors related to thermal stress and inflammatory processes [17]. Other benefits described for laying hens consists of increasing feed rates and productivity [18],[19],[20], and improvement of egg quality (reduced level of yolk cholesterol, increased shell thickness and higher egg weight [8](Chaucheyras Durand & Durand, 2009)) (Chung et al., 2015; Kurtoglu et al., 2004; Mazanko et al., 2017; Tang et al., 2015; Xu et al., 2006).
However, beneficial effects of probiotics have not only been described for poultry, and according to ReportLinker1 (2017), the probiotics market for animal feed has a projected value of USD 1,746.5 billion by the year 2022.
Lactic acid bacteria, especially Gram positive bacteria of the genus Lactobacillus, possess a high probiotic potential, and naturally inhabit the gut of mammals and birds [21],[22]. Nevertheless, lactobacilli make up less than 1% of the chicken GM [23]. Thus, the main objective of this work consisted in the design and implementation of a bioprospecting protocol for the isolation of lactobacilli from poultry feces, based on morphological and molecular identification, that could increase Lactobacillus isolation percentage compared to its representation in the GM. The protocol ends in an initial characterization of the probiotic potential inherent to the isolates obtained using antagonism tests.
The composition of the GM of chickens that inhabit in conditions which allow foraging behavior is different from the GM of chickens that are born in industrial conditions, confined in cages and under a traditional feed. Likewise, the varied diet of foraging chickens imply a healthier and more diverse gut ecosystem that may grant protection against certain pathogens (as the dietary effects on GM that have been observed in humans [24]. Therefore, the chickens that provided the samples live in artisanal production conditions that allow foraging behavior, which provides a varied diet of invertebrates, human food, corn, seeds, fruit, etc., without being restricted to a small cage.
Gram stain is a widely used technique for morphological identification of bacteria. However, to achieve species-level identification more sophisticated tools are required. MALDI-TOF mass spectrometry (MALDI-TOF MS) provides a quick and precise technique for bacterial identification that has been lately used in clinical laboratories around the globe [25]. It is based on laser evaporation of colonies, which generates volatile particles that are sucked by vacuum, with different time flights, into a detector, creating a spectrum unique to each species of bacteria. Another robust method used to identify bacteria is the sequencing of the 16S rRNA gene, a conserved locus whose sequence can be subjected to homology analysis for species assignment [26].
Antagonism tests are a common way to assess the probiotic potential of bacterial isolates. These tests are in vitro assays where interacting bacteria exhibit behavioral changes. Particularly, antagonism tests can provide information on the inhibitory effects of isolates against indicator organisms (e.g. pathogenic bacteria to be controlled), thus allowing the selection of promising isolates[27].
The importance of probiotics research aimed for poultry industry lays on the increasing consumption rates of poultry products, and therefore, the need to improve product quality and reduce costs. This need is especially conspicuous in Colombia, where according to the National Federation of Colombian Poultry Farmers (FENAVI, according to its Spanish acronym), the annual consumption of poultry meat per person went from 14.2 kgs in the year 2000, to 32.3 kgs for 2017, doubling in less than 20 years. During the same period, egg consumption per person has increased by 111 units/year.
2. Materials and methods
2.1 Sample processing
Under the collection framework permit granted by the National Environment Licensing Authority (in Spanish, Autoridad Nacional de Licencias Ambientales or ANLA), fecal samples were collected in three Colombian municipalities (Bahía Solano, Chocó: n = 10; Itagüí, Antioquia: n = 30; and Rionegro, Antioquia: n = 14), from chickens that inhabited in artisanal production conditions, which allowed foraging behavior. Samples were stored in Falcon tubes, and were transported in fridges. One gram of each sample was diluted fourfold in sterile water, and each dilution was then cultured on Merck’s ® solid MRS selective medium, which has a low pH and certain nutrients that benefit lactic acid bacteria growth.
2.2 Identiftcation
The isolates that grew on selective medium were identified by three methods. First, a Gram stain was used to select colonies that met the morphologic characteristics of lactobacilli (lengthened Gram positive bacilli).
The colonies selected by Gram stain were then identified by a bioMérieux® MALDI-TOF MS spectrometer with a 337 nm nitrogen laser (fixed focus) (VITEK® MS). Colonies were identified based on spectra made of protein flight times, whose weight ranged between 1 and 500 kDa. This range includes ribosomal and some housekeeping proteins. Spectra were compared with the VITEK® MS V3.2.0 CE-marked database, using MYLA® middleware software, to obtain identity of colonies. High fidelity identifications were preserved at -70 oC in Merck’s ® MRS medium with 20% of glycerol.
To confirm MALDI-TOF MS identification, eight isolates representing all of the Lactobacillus species found by mass spectrometry, were randomly selected. For each colony, a total DNA extraction and PCR amplification of the 16S rRNA gene were performed.
DNA was extracted using Ultra Clean ® microbial DNA isolation kit (MO BIO laboratories, inc). DNA was amplified via PCR using Taq Polymerase, recombinant (5 U/L) (Thermo Fisher Scientific, inc). Primers: 27F (AGAGTTTGATCMTGGCTCAG) and 907R (CCGTCAATTCMTTTRAGTTT) (Lane, 1991). The obtained amplicons were sequenced by Sanger method, and consensus sequences were created using Geneious ®. To determine the isolate species, 16S rRNA gene sequences were aligned to NCBI database using BLAST [28].
2.3. Antagonism tests
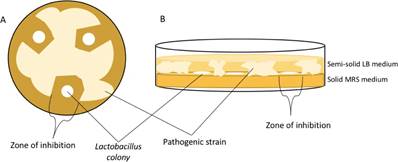
Antagonism test were performed in order to initially assess the probiotic potential of the same isolates selected for 16S rRNA gene identification. These were tested against five pathogenic strains provided by The Universidad EAFIT strain collection: Staphylococcus aureus, Enterococcus faecium, Candida albicans, Pseudomonas spp., and Salmonella spp. The eight isolates were cultured in Petri dishes, on solid MRS Merck ® medium along a negative control strain Lactobacillus reuteri (ATCC® 23272). 24 hours later, the five pathogenic strains were inoculated in 1% semi-solid LB broth (lysogenic broth, Merck ® ) which was then poured over the MRS solid medium, with already visible lactobacillus colonies that grew overnight. The Petri dishes containing both growth media were incubated at 37 oC for 24 hours. After this incubation period, the presence of inhibition zones was visually established according to the presence or absence of pathogen growth surrounding the vicinity of lactobacillus colonies [Figure S1].
3 Results and discussion
3.1. Identifted isolates
114 colonies were obtained from the initial cultures of the diluted fecal samples, and after Gram stain, 49 of the 114 original colonies met the morphologic characteristics of lactobacilli. These 49 colonies were submitted to MALDI-TOF MS, and by this method, 31 were identified with 99% precision. The 31 isolates belonged to the order Lactobacillales, distributed in three families (Enterococcaceae, Streptococcaceae, and Lactobacillaceae), four genera (Enterococcus, Pediococcus, Streptococcus, and Lactobacillus), and in eight species of bacteria [Figure 1]. 24 isolates belonged to the genus Lactobacillus were distributed in four species [Figure 1], which accounts for 77.4% of the 31 isolates identified by mass spectrometry and 21% of the original 114 colonies. This suggests an effective selection by the MRS medium, and by morphological selection using Gram stain, given the fact that the majority of isolates belonged to the genus Lactobacillus, and all of them are grouped under the order Lactobacillales and the family Lactobacillaceae (lactic acid, non-sporulating, Gram positive bacteria [29], fulfilling the necessary physiological and morphological characteristics in order to grow in MRS medium and be selected by Gram stain. Furthermore, the family Lactobacillaceae represents less than 1% of the total microbial population of the chicken gut [23] and our bioprospecting protocol was able to augment Lactobacillaceae representation up to 21% of the fecal samples.
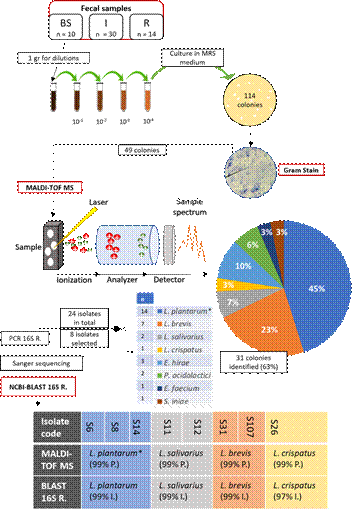
Figure 1 Identification process and results. Fecal samples were collected on three locations with different sample size: Bahia Solano (BS), Itagüi (I), Rionegro (R). After four dilutions, 114 colonies grew in selective medium. 49 were singled out by Gram stain for further identification by MALDI-TOF MS. This method identified the species of 31 (63%) isolates with 99% precision (P.), with the exception of 14 isolates asigned to a group of three closely related species, Lactobacillus plantarum/paraplantarum/pentosus (*). To confirm MS identification, NCBI-BLAST of 16S rRNA gene was performed with >97% identity results matching MALDI-TOF MS identification.
The exact species for 14 of the 24 Lactobacillus isolates couldn’t be identified by MALDI-TOF MS. These 14 isolates were assigned to a group of three closely related species, Lactobacillus plantarum/paraplantarum/pentosus. In order to confirm MALDI-TOF MS results, eight isolates representing the four lactobacillus species previously identified (including Lactobacillus plantarum/paraplantarum/pentosus) were randomly selected for 16S rRNA gene identification. All NCBI-BLAST results of the gene sequences confirmed MALDI-TOF MS identification [Figure 1]. Furthermore, within the eight isolates selected, three of them grouped by mass spectrometry in L. plantarum/paraplantarum/pentosus cluster (S6, S8, and S14) were identified with 99% of identity (E-value < 0.1) as L. plantarum, by majority of matches with 16S rRNA gene sequences BLAST.
3.2 Antagonism tests
Table 1 summarizes antagonism tests results between eight Lactobacillus, previously selected for 16S rRNA gene identification, and five pathogenic strains that are commonly found in the gut of chickens.
Table 1 Antagonism tests. Eight Lactobacillus isolates tested against five pathogenic strains. +: inhibition zone present; : inhibition zone absent; EF: E. faecium; CA: C. albicans; S: Salmonella spp.; SA: S. aureus; P: Pseudomonas spp.
| Species | Code | EF | CA | S | SA | P |
| L. plantarum | S6 | + | - | - | - | + |
| L. plantarum | S8 | - | - | + | - | - |
| L. plantarum | S14 | - | - | + | - | - |
| L. brevis | S31 | - | - | - | - | - |
| L. brevis | S107 | - | - | - | - | - |
| L. salivarius | S11 | + | + | + | + | + |
| L. salivarius | S12 | + | + | - | + | + |
| L. crispatus | S26 | - | - | - | - | - |
Five of the isolates selected, three corresponding to L. plantarum, and two identified as L. salivarius, showed a degree of inhibitory effect against at least one pathogenic strain. S11 and S12, the two L. salivarius isolates, showed the broadest spectrum of inhibition, hindering the growth of all pathogenic strains (S11) or of all with the exception of Salmonella spp. (S12). In agreement with these results, L. salivarius anti-infective ability has already been proved in mice against Listeria monocytogenes [30], against Salmonella enteritidis infections in chickens (Pascual et al., 1999), hindering of biofilm formation by C. albicans in children with caries (krzysciak et al., 2017), in vitro growth inhibition of Helicobacter pylori [31], and other bacteria belonging to the genera Bacillus, Enterococcus, Staphylococcus, Clostridium, Pseudomonas, Streptococcus, Escherichia, and Bifidobacterium [32].
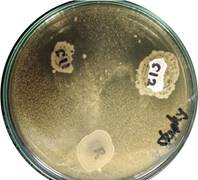
Figure 2 Inhibition of S. aureus growth surrounding L. salivarius colonies (S11 and S12). No inhibition surrounded L. reuteri ATCC ® 23272 control colony (R)
Our results suggest S11 and S12 ability to produce either one type of broad spectrum antimicrobial compound, or several types of compounds that inhibit the growth of the variety of pathogen strains tested, all markedly different: S. aureus [Figure 2] and E. faecium (Gram positive), Salmonella spp. and Pseudomonas spp. (Gram negative), in addition to a yeast (C. albicans). One possible explanation for S11 and S12 broad spectrum inhibitory activity is production of several types of bacteriocin (selective broad spectrum antimicrobial peptides) [33].
3 Conclusions
For this study, we designed a precise and efficient bioprospecting protocol directed to lactobacilli isolation from poultry feces. Based on four techniques of filtering and identification of colonies (in order: selective medium, Gram stain, MALDI-TOF MS, and 16S rRNA gen sequence BLAST), this protocol allowed the robust identification of 24 Lactobacillus isolates and four species (along with other four lactic acid bacteria species) from an initial sample of 114 colonies that grew on MRS selective medium. Thus, with a rate of 21% (24/114) isolation, this protocol highly increases Lactobacillus yield compared to its representation of less than 1% in the chicken GM (21-fold).
The protocol ends with a preliminary assessment of a possible probiotic potential of the isolates using antagonism tests, and our results showed positive for five isolates belonging to two Lactobacillus species, meaning that these isolates inhibited the growth of at least one pathogen strain tested. From these results a probiotic potential hierarchy can be proposed: with S11 and S12 in the first and second place, respectively (L. salivarius isolates); followed by S6 in third place, with S8 and S14 sharing the fourth place (L. plantarum isolates); finally, sharing the last place are S31, S107 (L. brevis isolates), and S26, L. crispatus single isolate. Positive antagonism results could also be increased by performing tests to a larger sample of isolates and pathogen strains.
Given the superior inhibition ability observed for L. salivarius isolates, we propose that these bacteria could produce either a wider variety of antimicrobial substances or a single molecule with a broad antimicrobial spectrum, and that this production often reaches the minimal inhibitory concentration (MIC). On the other hand, we suggest several hypothesis that explain that L. brevis and L. crispatus isolates did not show antagonistic properties: it is possible that both isolates produce antimicrobial compounds but their quantities do not reach the MIC; these isolates lost the genetic potential to produce antimicrobial compounds; antimicrobial compounds are produced but they are not effective against the pathogenic strains tested; growth conditions hinder expression of antimicrobial compounds.
A way to confirm these hypotheses is by directly analyzing antimicrobial production by a quantification technique (e.g. HPLC) or indirectly by a transcriptomic analysis directed to the identification of transcripts associated with antimicrobial compounds. Bacteriocin production could be confirmed by quantifying the sensitivity of protease activity, or by a proteomics approach that would allow identification of the different types of bacteriocins produced.
The probiotic potential assessment done in this study is preliminary, however, due to the reduced number of isolates and pathogen strains tested, as well as the constant conditions in which these tests were conducted (pH, salts, and temperature). Furthermore, pathogen inhibition in a Petri dish is just one of the measurable characteristics that imply benefits to the host of the probiotic bacterium. Inhibition of pathogen adhesion to gut epithelia [34], host immune system induction [35], influence on nutrients uptake [36], and resistance of the probiotic bacterium to bile salts and low pH [37], are just some characteristics that make up a high potential probiotic. Then, to achieve a more holistic probiotic potential characterization, it is required to measure several parameters that take into account the mentioned properties in vitro and in vivo.