Introduction
World farmed shrimp production volume decreased significantly in 2013 as a result of early mortality syndrome in some countries in Asia and Latin America (FAO, 2014). Therefore, efforts have been made to develop hyper-intensive crops through biofloc technology (Emerenciano et al., 2013). The biofloc is a complex of living organisms that are closely associated with particulate organic matter and are maintained in suspension by continuous aeration. Plankton and bacteria, which are part of this complex, metabolize nitrogenous waste excreted by fed-farmed animals. This property of biofloc is due to its conformation and is being used worldwide for culturing various aquatic animal species because this system maintains good water quality in densely populated systems (Avnimelech, 2012). In Mexico, some farmers have seen that biofloc has a preventive and even healing capacity against bacterial diseases. It is known that biofloc promotes immunological and antioxidant responses in different shrimp species (Xu and Pan, 2014; Cardona et al., 2015). Shrimp immune system depends mainly on innate immunity and is divided in humoral and cellular immunity (Lee and Söderhäll, 2002; Cerenius and Söderhäll, 2004; Jiravanichpaisal et al., 2006). In crustaceans, innate immunity recognizes invading microorganisms and triggers various defense mechanisms to eliminate pathogens (Söderhäll and Cerenius, 1992). The pathway through which cells identify microorganisms is through pattern recognition receptors (PRRs). This PRRs regulate the expression of down-stream immune-related genes after stimulation by pathogens associated molecular patterns (PAMPs), such as lipopolysaccharide (LPS), β-I,3-glucans, and peptidoglycans (Wang and Wang 2013; Chen et al., 2016).
Hence, our hypothesis is that the microorganisms present in a biofloc system (BF) may trigger the immune response of shrimp. In the present study, post-larvae (PL) shrimp were exposed to a RAS with a mature heterotrophic BF or without BF (no-BF), and the transcriptional response of seven immune-related genes was monitored.
Materials and methods
Ethical considerations
This study complies with the Mexican official Standard NOM-0062-ZOO-1999, technical specifications for production, care and use of laboratory animals.
Experimental culture systems
Two identical RAS (800 L each) were designed. Each system consisted of the main tank with 400 L of seawater, pumped to an elevated tank of 100 L. The water was distributed among 6 glass aquariums (50 L each) by gravity (~50 L/h). The aquariums were connected to a return tube to the main tank. Every container was supplied with vigorous aeration using air stones in aquariums and aero-tube in the main tank. Bunches of polypropylene fibers (50 g/L) (Brand COVERPACK-Mexico) were placed into the main tank of RAS no-BF, as a substrate for a biological filter.
Juvenile shrimp (1 ± 0.5 g = 30 per aquarium) were placed in the system, and cane molasses was added as a source of organic carbon (0.2 g/L daily) to promote biofloc formation. When the BF reached the appropriate parameters (Table 2), the juvenile shrimp were removed from the aquariums for the transcriptional response experiment.
Experimental design
One batch of 2,000 PL (mean body weight of 150 ± 30 mg) was acquired from a commercial hatchery and acclimatized for 24 h at 35 g L-1 salinity before being placed in the experimental systems.
After removing juvenile shrimp from experimental aquariums (above), 150 PL shrimp were stocked into each aquarium. At this time, we collected six PL shrimp to evaluate the original expression (T0). Later, one PL shrimp per replicate (six per treatment) was collected at different times (3, 6, 9, 12, 18, 24, 36, 48, and 72 h). Each PL shrimp was placed individually in Trizol reagent for RNA extraction and stored at -70 °C until analyses.
Physicochemical parameters
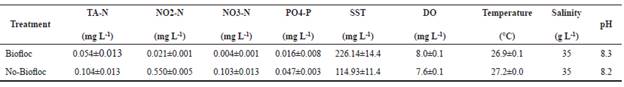
Prior to the experiment, pH, water salinity, temperature (°C), and dissolved oxygen (DO; mg L-1) were measured daily in each aquarium of both systems. Total ammonia (mg L-1), nitrite (mg L-1), nitrate (mg L-1), phosphate (mg L-1), alkalinity (mg CaCO3 L-1), and total suspended solids (TSS, mg L-1) were analyzed twice a week until suitable levels for a biofloc system were reached. The chemical parameters were analyzed following the methods described by Strikland and Parsons (1972) and APHA (1998).
Bacterial analyses
Water samples for bacterial analyses were collected from each aquarium following standard procedures (APHA, 1998; Gómez-Gil, 2006). Presumptive Vibrio sp.was detected in a plate per triplicate with Thiosulfate-citrate-bile salts-sucrose agar (TCBS agar; Difco, Detroit, MI, USA) supplemented with 2.5% NaCl and incubated for 24 h at 30 °C before counting (CFU mL-1). Presumptive Bacillus sp. were determined. Briefly, water samples were incubated at 80 °C for 10 min to debug the sample of non-spore forming bacteria. Subsequently, Bacillus sp were detected in a plate per triplicate with trypticase soy agar (TSA, BD, Bioxon, Sparks, MD, USA) supplemented with 2% NaCl and incubated for 24 h at 37 °C before counting.
RNA extraction and cDNA synthesis
Total RNA was extracted from individual PL using 1 mL of TRI REAGENT (Molecular Research Center, Inc. Cincinnati, OH, USA) following manufacturer’s instructions and washing RNA pellet twice before drying. Total RNA was quantified on a NanoPhotometer (Pearl, Implen®, Westlake Village, CA, USA) considering the absorbance ratio 260/280 nm between 1.8 and 2.0 as acceptable purity range.
To transcribe the cDNA, an aliquot of the to tal RNA was diluted at 300 ng/μL (25 μL) and then treated with DNAse I (1 U/μL, Sigma-Al drich, St. Louis, MO, USA) following manufac turer’s instructions.
The cDNA synthesis was performed with Im prom II (Promega, Madison, WI, USA) and the oligo (dT20) primer using 1.0 μg of RNA, fol lowing manufacturer’s instructions. The cDNA was diluted 10 times with ultrapure water and stored at -70 °C until qPCR analysis and 2.5 μL of this cDNA dilution was used as template in each qRT-PCR reaction.
Primer design and detection of ADP ribosylation factor 4 in Litopenaeus vannamei
Arf4 primers were designed from Marsupenaeus japonicus (GeneBank accession GQ279375) and Penaeus monodon (GeneBank accession KM210090) to amplify the full sequence of LvArf4 (543 bp). The PCR products were resolved on a 1.5% agarose gel. The bands were excised from the gel and purified with GENECLEAN Kit (MP Biomedicals, Santa Ana, CA, USA), cloned in the pGEM-T Easy Vector System (Promega), and sequenced by GENEWIZ (South Plainfield, NJ, USA).
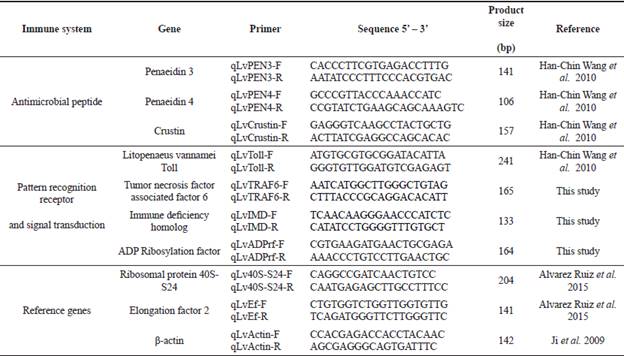
Specific primers for RT-qPCR were designed from the confirmed sequences (Table 1).
Quantitative real-time PCR (RT-qPCR) analysis
Relative expression was measured by RT-qPCR with specific primers (Table 1) using a CFX96 Real-Time PCR thermal cycler (Bio- Rad Laboratories, Hercules, CA, USA). The expression of target genes was normalized with the geometric mean of 40S-S24, Ef-2, and β-actin (Table 1). The stability of reference genes, Cq transformation, PCR master mix, efficiency, and amplification conditions were done as described by Alvarez-Ruiz et al. (2015).
Statistical analyses
Up-regulated expression was determined by statistical analysis using a two-way ANOVA. Expressions higher than the control group (T0) were considered as up-regulated. Statistical differences in the bacterial population between treatments were done by Student’s t-test. The analyses were conducted using STATISTICA package v6 (StatSoft, Tulsa, OK, USA). Statistical significance was set at (p<0.05).
Water quality and bacterial population
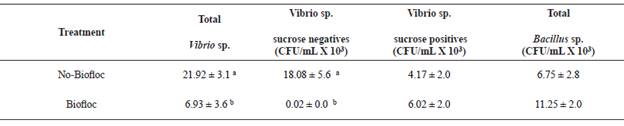
The water quality parameters were within acceptable ranges for shrimp growth at the beginning of the experiment (Table 2). Total Vibrio sp. count presented significantly higher values in no-BF than in BF (21.9 × 103 CFU vs. 6.9 × 103 CFU).
There were no sucrose-negative colonies in BF, hence, presenting significant differences in regard to no-BF (18.1 × 103 CFU). There were no differences in sucrose-positive colonies between BF and no-BF (6.0 × 103 CFU vs. 4.2 x 103 CFU) or in populations of Bacillus between BF and no-BF (6.8 × 103 CFU vs. 11.3 × 103 CFU) (Table 3).
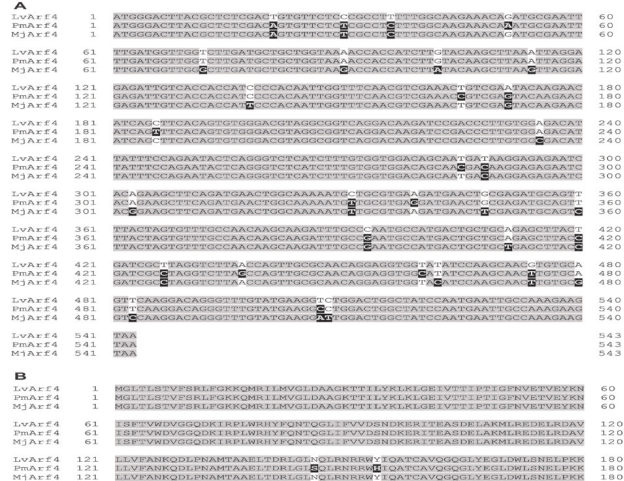
Figure 1 Nucleotide (A) and deduced amino acid (B) sequences alignment of the ADP-ribosylation factor 4 of L.vannamei (LvArf4) from Mexico compared to the corresponding Arf4 sequences from M.japonicus and P.monodon. Grey shade indicates identical nucleotides or amino acids in all species (conserved region). Dark shade indicates differences as compared to L.vannamei sequence.
Cloning and sequence analyses of ADP ribosylation factor 4
The full sequence of the ADP ribosylation factor 4 in L.vannamei was sequenced (GenBank ACCESSION MK471369). Blast analyses showed 95 and 97% nucleotide identity with the Arf4 from M.japonicus and P.monodon, respectively (Figure 1A).
The predicted amino acid sequence of LvArf4 of L.vannamei showed 100 and 99% identity to MjArf4 and PmArf4, respectively (Figure 1B).
Transcriptional response of shrimp immune-related genes
Results are grouped below according to the gene role in the shrimp defense mechanism.
Antimicrobial peptides (Pen3, Pen4, crustin).
Pen3 gene was up-regulated at 6 and 24 h in BF, and 3 and 18 h in no-BF. The expression level of Pen3 in no-BF was higher than in BF at 3 h, but lower at 6 h (Figure 2A).
Pen4 gene was up-regulated at 3 h in no-BF and 6 h in BF. Besides, the expression of Pen4 in no-BF was significantly higher than in the BF at 3 and 48 h (Figure 2B).
Crustin gene was up-regulated in both systems at 3 and 6 h. The expression of crustin in the BF was significantly higher than in no-BF at 36 h (Figure 2C).
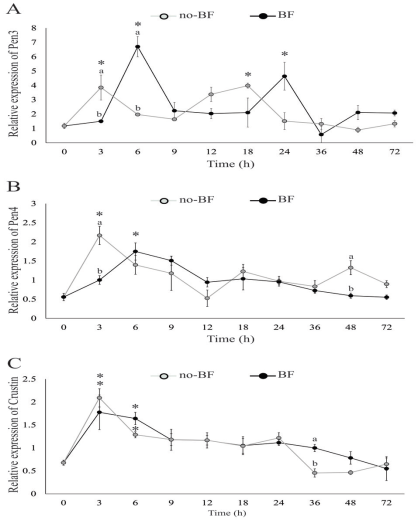
Figure 2 Relative expression of genes related to antimicrobial peptides [Pen3 (A), Pen4 (B) and crustin (C)] in post-larvae after being placed into RAS with biofloc (BF) or without biofloc (no-BF). Up-regulated expression is marked with an asterisk (*). Significant differences between treatments are represented by different letters (p<0.05).
Pattern Recognition receptor and signal transduction (LvToll TRAF6, IMD, Arf4).
Upon placing shrimp in the cultures, LvToll was up-regulated at 3 h in no-BF, and at 12, 36, and 48 h in BF. The expression of LvToll was higher in BF than in no-BF at 36 and 48 h (Figure 3A).
LvTRAF6 expression remained stable in BF. However, LvTRAF6 was up-regulated at 3 h in no-BF, and was higher than in BF. Contrarily, the expression of TRAF6 was higher in BF at 72 h (Figure 3B).
IMD expression was stable throughout the experiment in both BF and no-BF. However, IMD expression was higher in BF than in no-BF at 3 h (Figure 3C).
Arf4 expression was not up-regulated throughout the experiment. However, the transcription of Arf4 in BF was significantly higher than in no-BF from 36 to 72 h (Figure 3D).
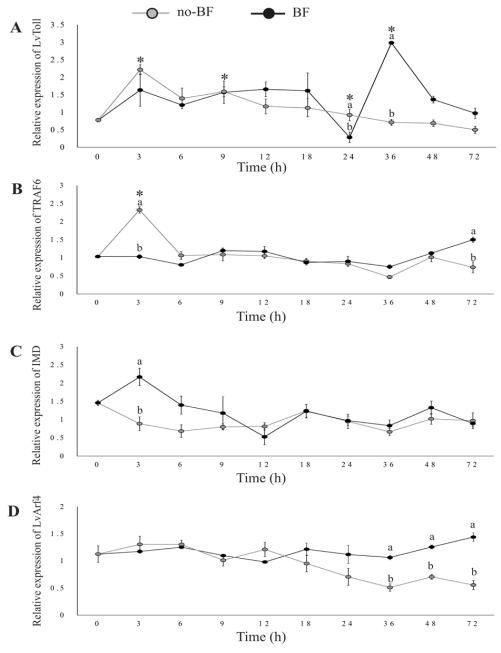
Figure 3 Relative expression of genes related to pattern recognition receptor and signal transduction [L.vannamei Toll (A), tumor necrosis factor receptor-associated factor 6 (B), immune deficiency homolog (C), and L.vannamei ADP-ribosylation factor 4 (D)]. Experimental conditions and data presentation are the same as for Figure 2.
Discussion
Our understanding of the innate immune response of shrimp, when cultured under biofloc conditions, is still in its early stages. Besides, the RAS-no-BF for rearing shrimp has shown similar productive capacity to the biofloc systems (Esparza-Leal et al., 2015; Ray and Lotz, 2017). Hence, in this study, we evaluated whether the heterotrophic micro-ecosystem (biofloc system) and RAS-no-BF promote the transcription of immune genes in L.vannamei.
We studied the mRNA levels of seven genes through real-time PCR, including ADP-ribosylation factor 4 from L.vannamei, which, to the best of our knowledge, is reported for the first time in this study.
Water quality
At the beginning of the bioassay, the physicochemical parameters were kept within acceptable ranges for rearing L.vannamei in BF and no-BF cultures (Esparza-Leal et al., 2015; Ray and Lotz, 2017; Schveitzer et al., 2018).
Detection of ADP-ribosylation factor 4 in L.vannamei (LvArf4)
An Arf gene has been described for the first time in shrimp (M.japonicus), and showed a delayed transcriptional response against WSSV infection (24, 48, 72, and 96 h postinfection) (Zhang et al., 2010). Although we lacked information about the Arf4 sequence in L.vannamei, we were able to detect, clone, and sequence for the first time the Arf of L.vannamei. Sequence identity was 95% with MjArf4 sequence (Zhang et al., 2010) and 97% with PmArf4. This result indicates that Arf4 is a conserved protein among shrimp species.
Expression of antimicrobial peptides
AMPs constitute vital components of the innate immune system of shrimp, acting as immune effectors by killing or inhibiting the growth of their microbial targets (Destoumieux et al., 1997). Penaeidins are members of a family of antimicrobial peptides that have been proven to be a rapid response mechanism to resist microbial invasions (Destoumieux et al., 2000). Crustin is an antibacterial peptide that has antimicrobial activity against Gram-positive marine bacteria (Jiang et al., 2015). However, recently, an up-regulation of crustin transcripts has been reported after challenging with V.parahaemolyticus and WSSV (Rubio-Castro et al., 2016).
We registered an up-regulation of Pen3, Pen4, and crustin transcripts in shrimp during the first hours after contact with BF or no- BF, suggesting that the shock between the shrimp internal bacterial population and the microorganisms present in the culture systems promotes the synthesis of AMPs. Since AMPs are stored in shrimp granulocytes, and released after microbial challenge (Destoumieux et al., 2000), the up-regulation recorded in the BF could be an indicator of immune-competence promoted by the microorganisms living in the flocs. On the other hand, the microbial population present in the biological filter could explain the overexpression recorded in no-BF.
Differences in the effectiveness of inhibition have been observed between Pen3 and Pen4 for several species of Bacillus and Micrococcus (Cuthbertson et al., 2008). In the present study, we found a population of Bacillus sp. in both systems, BF and no-BF, and registered an up-regulation of the three AMPs between 3 and 6 h. Besides, Pen3 showed up-regulation at different times in both treatments. This suggests that Pen3 is more sensitive to the microbial population in the culture systems.
Expression of pattern recognition receptor and signal transduction
There are several ways in which shrimp cells respond against pathogens or foreign molecules; this includes Toll-like receptors. The expression of the Toll gene in shrimp is stimulated by PAMPs and, thus, induces a response against them (Liu et al., 2016).
Our results showed a sudden up-regulation at 3 h in no-BF, perhaps as a response to the microorganisms in the water. On the other hand, a remarkable up-regulation was registered at 12, 36, and 48 h in the BF; this expression was higher than in no-BF at 36 and 48 h, suggesting that after a period of adaptation the LvToll expression is modulated by the culture conditions in the biofloc.
The TRAF6 is one of the key adaptor molecules in the Toll-like receptor signal transduction that triggers downstream cascades involved in innate immunity (Kobayashi et al., 2004). It has been documented that transcription of Toll and TRAF6 genes responds positively to a WSSV infection in a primary hemocyte culture of P.monodon (Deepika et al., 2014). In contrast, TRAF6 on shrimp is down-regulated in the intestine after WSSV or Vibrio alginolyticus infection (Wang et al., 2011). In the present study, we recorded a sudden up-regulation of TRAF6 at 3 h in no-BF, but not in BF. These findings suggest that Toll and TRAF6 play a role in the immune response of shrimp, which may be regulated by pathogens of different etiology or by the microbial community residing in the culture system.
In addition, LvIMD transcription responds to a WSSV infection and can induce expression of Pen4 (Wang et al., 2009). Besides, it is involved in regulating some AMP genes like crustin 1 and 3, anti-lipopolysaccharide factor 6 and 8, and lysozyme 2 (Lan et al., 2013). In this study, transcription of LvIMD remained stable throughout the experiment in both BF and no-BF treatments and, at the same time, we recorded an up-regulation of Pen3, Pen4, and crustin. This indicates that regulation of AMPs in the treatments was not promoted by IMD. Instead, the recorded expression of LvToll and TRAF6 genes suggests that the transcription of AMPs was probably promoted by genes related to the Toll-like receptors pathway. In this sense, Lan et al. (2013) evaluated Feneropenaeus chinensis IMD transcription after a challenge against Vibrio anguillarum and did not find marked differences in hemocytes and no differences in the gill. In contrast, Liu et al. (2016) showed a pronounced transcriptional response of LvIMD in organisms challenged against Gram-negative or Gram-positive bacteria.
Taken together, these findings evidence that further investigations focused on demonstrating how the IMD pathway is related to the innate immune system of the shrimp are necessary.
ADP ribosylation factors (Arfs) are a family of small guanine nucleotide binding proteins that control membrane traffic and organelle structure and have essential functions, including the recruitment of coat proteins that promote sorting of cargo molecules into vesicles (Donaldson and Jackson, 2011). The transcription of LvArf4 showed minor differential expression between no-BF and BF at 36, 48, and 72 h. These results suggest that the BF promotes a differential regulation of the LvArf4 gene at medium term; a more detailed analysis of this gene is necessary.
In conclusion, the transcriptional response of immune-related genes induced by a RAS with BF and no-BF occurs as an early response, possibly as a result of the shock with the bacterial community attached to flocs or the biological filter of no-BF. It seems that the biological filter in the RAS no-BF can maintain a bacterial population that promotes a similar response to that elicited in the BF. Whether this differential transcriptional response confers immunity to shrimps against pathogens is something that remains to be further investigated.