Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Colombiana de Ciencias Pecuarias
Print version ISSN 0120-0690On-line version ISSN 2256-2958
Rev Colom Cienc Pecua vol.23 no.3 Medellín July/Sept. 2010
Genetic characterization of the Hartón del Valle, Angus, Brangus, Holstein, and Senepol cattle breeds in Colombia, using ten microsatellite markers¤
Caracterización genética de las razas Hartón del Valle, Angus, Brangus, Holstein y Senepol en Colombia, usando 10 marcadores microsatélites
Caracterização genética das raças Hartón del Valle, Angus, Brangus, Holandês e Senepol na Colômbia, usando 10 marcadores microsatélites
Alba E Montoya1,2, Biol, MSc; Mario F Cerón-Muñoz1,2*, Zoot, PhD; Manuel A Moreno1,3, Biol, PhDc; Edwin Martínez1, Est Biol; Juan D Corrales1, Zoot, est MSc; Juan F Tirado1, Zoot; Samir J Calvo1, Zoot.
1 Grupo de Genética, Mejoramiento y Modelación Animal (GaMMA), Instituto de Biología, Facultad de Ciencias Agrarias, Universidad de Antioquia, Medellín, Colombia. 2 Docente, Facultad de Ciencias Agrarias, Universidad de Antioquia, AA 1226, Medellín, Colombia. 3 Docente, Instituto de Biología, Universidad de Antioquia, AA 1226, Medellín, Colombia.
(Recibido: 5 septiembre, 2009; aceptado: 18 mayo, 2010)
Summary
The objective of this paper is to establish a genetic characterization of the Senepol (S, n=49), Holstein (H, n= 60), Hartón del Valle (HV, n=60), Angus (A, n=61) and Brangus (Br, n=60) cattle breeds in Colombia, by using the following microsatellite markers: SPS115, INRA64, ETH225, ETH10, BM1824, INRA37, TGLA122, TGLA126, INRA32, and BM2113. A total of 142 alleles were obtained for ten analyzed loci, considering the five cattle breeds as a whole. The number of alleles per locus ranged from 9 (INRA64 and 1824) to 22 (TGLA122). The expected heterozygosity was between 0.79 (INRA32) and 0.90 (INRA37) in all the cattle breeds, respectively; and medium heterozygosity was 0.84. The average number of alleles per breed varied from 9.2 in the Senepol breed to 10.3 in the Holstein breed. The expected heterozygosity range varied from 0.75 in the Hartón del Valle breed and 0.82 in the Holstein breed, with an average of 0.79. Hardy Wienberg disequilibrium was observed (p>0.05) when the populations were analyzed with all the markers. All the populations presented a heterozygote deficit, which could be the result of a strong endogamy tendency within all the herds. The markers used in this study allowed a genetic characterization of the analyzed populations. The microsatellites panel in the Hartón del Valle breed should be increased in order to increase the reliability value. Microsatellite panels could solve parenthood cases for the remainder breeds.
Key words: cattle, genetic variability, molecular marker.
Resumen
El objetivo de este trabajo fue caracterizar genéticamente las razas bovinas Senepol (S, n=49), Holstein (H, n= 60), Hartón del Valle (HV, n=60), Angus (A, n=61) y Brangus (Br, n=60) en Colombia, con los marcadores microsatélites SPS115, INRA64, ETH225, ETH10, BM1824, INRA37, TGLA122, TGLA126, INRA32 y BM2113. En total, 142 alelos fueron encontrados en los diez loci analizados, considerando las cinco razas como un todo. El número de alelos por locus estuvo entre 9 (INRA64 y BM1824) y 22 (TGLA122). La Heterocigosidad esperada a través de todas las razas varió entre 0.79 (INRA32) y 0,90 (INRA37) y heterocigosidad media esperada de 0.84. El número promedio de alelos por raza varió de 9.2 en la raza S a 10.3 en la raza H. El rango de la Heterocigosidad esperada entre las razas varió entre 0.75 en la raza HV y 0.82 en la raza H, con una media de 0.79. Al analizar las poblaciones con el total de marcadores, todas se encontraron en desequilibrio de Hardy Weinberg (p>0.05). Todas las poblaciones presentaron un déficit de heterocigotos, para todas las poblaciones, lo que podría ser el resultado de la fuerte tendencia a la endogamia dentro de los diferentes hatos. Los resultados indicaron que los marcadores utilizados en este estudio permitieron caracterizar genéticamente las poblaciones analizadas. En el caso de la Raza HV, se debe aumentar el panel de microsatélites para aumentar el valor de confiabilidad. Para las demás razas el panel de microsatélites permitiría resolver casos de filiación.
Palabras clave: ganado, marcadores moleculares, variabilidad genética.
Resumo
O objetivo do presente trabalho foi caracterizar geneticamente as raças Senepol (S, n=49), Holandês (H, n= 60), Hartón del Valle (HV, n=60), Angus (A, n=61) e Brangus (Br, n=60) na Colômbia, com os marcadores microsatélites SPS115, INRA64, ETH225, ETH10, BM1824, INRA37, TGLA122, TGLA126, INRA32 e BM2113. Em total, 142 alelos foram encontrados nos 10 satélites analisados nas cinco raças. O número de alelos esteve entre 9 (INRA64 e BM1824) e 22 (TGLA122). A heterocigosidade esperada a través de todas as raças variou entre 0.79 (INRA32) e 0.90 (INRA37) e heterocigosidade esperada de 0.84. O número médio de alelos por raça variou de 9.2 na raça S a 10.3 na raça H. O rango de heterocigosidade esperada entre raças variou entre 0.75 na raça HV e 0.82 na raça H, com una media de 0.79. Ao analisar as populações com o total de marcadores encontraram-se o desequilíbrio Hardy Weinberg (p>0.05). Todas as populações apresentaram um déficit de heterocigotos, o que poderia ser o resultado da forte tendência de endogamia nos diferentes rebanhos analisados. Os resultados indicaram que os marcadores utilizados em este estúdio permitiram caracterizar geneticamente as populações analisadas. No caso da raça HV deve-se aumentar o número de microsatélites para aumentar o valor de confiabilidade. Para as demais raças os microsatélites analisados permitiriam resolver casos de paternidade.
Palavras chave: gado, marcador molecular, varibilidade genética.
¤ Para citar este artículo: Montoya AE, Cerón-Muñoz MF, Moreno MA, Martínez E, Corrales JD, Tirado JF, Calvo SJ. Genetic characterization of the Hartón del Valle, Angus, Brangus, Holstein, and Senepol cattle breeds in Colombia, using ten microsatellite markers. Rev Colomb Cienc Pecu 2010; 23:283-291.
* Autor para correspondencia: Mario F Cerón-Muñoz. Escuela de Producción Agropecuaria. Facultad de Ciencias Agrarias, Universidad de Antioquia. AA1226, Medellín, Colombia. E-mail: mceronm@hotmail.es
Introduction
Livestock breeds have been the product of centuries of natural and artificial selection. Breeds have been selected to fit a wide range of environmental conditions and human needs. The selection of a few highly productive breeds has caused the decrease of many other non-productive breeds (Maudet et al., 2002).
In Colombia there is a large variety of native and foreign breeds; most of them have adapted to the diversity of geographical conditions of these country. Therefore, it is thought that these breeds present an enormous genetic diversity due to the fact that they come from different regions could explain their genetic variability.
Microsatellite markers (STR) have made possible the study in numerous breeds around de world, something like that. Currently, there are more than 1200 STR known in cattle (Kappes et al., 1997) some of them approved by the International Society of Animal Genetics (ISAG) and the Food and Agriculture Organization (FAO, 2007). The usefulness of microsatellite markers have been documented in bovine breeds for the estimation of genetic diversity (Ciampolini et al., 1995; Peelman et al., 1998; Zamorano et al., 1998b; Steigleder et al., 2004; Barrera et al., 2006a; Martínez et al., 2006) and the relationships among livestock breeds (Moreno et al., 2001; Hansen et al., 2002; Maudet et al., 2002; Radko et al., 2005; Barrera et al., 2006b), as well as genetic characterization (Zamorano et al., 1998a; Lara et al., 2005; Martínez et al., 2005; Calvo et al., 2009; Martínez et al., 2009), individual profiles, and paternity tests (Giovambattista et al., 2001; Mommens et al., 1998; Sanz et al., 2002) and genealogical-registries control (Jamieson, 1994).
In general, the distribution of genetic polymorphisms tends to be quite homogeneous among the different populations involved. Nevertheless, in the variability extend, specific characteristics for each population group are usually noted. (Barrera et al., 2006a; Barrera et al., 2006b, Calvo et al., 2009, Hansen et al., 2002; Martínez et al., 2009; Maudet et al., 2002; Mommens et al., 1998; Moreno et al., 2001; Radko et al., 2005; Steigleder et al., 2004; Zamorano et al., 1998a; Zamorano et al., 1998b). This could be achieved through the genetical characterization of the animals in each region.
This paper was conducted to study genetic diversity among the Hartón del Valle, Angus, Brangus, Holstein, and Senepol cattle breeds, all of them found in Colombia and well adapted to the diversity of its weather and geographical conditions, using ten microsatellite markers located in different chromosomes.
Materials and methods
Population Sample
Sample was constituted as follows: Senepol (S, n=49) from Hacienda Río Piedras, municipality of Jericó, Antioquia; Holstein (H, n= 60) from the municipality of San Pedro de los Milagros, Antioquia; Hartón del Valle (HV, n=60) from Hacienda Sanjonhondo, located in Tuluá municipality, Valle del Cauca; Angus (A, n=61) and Brangus (Br, n=60), both from Centro de Investigación en Biotecnología y Reproducción (Cibre) of the Universidad San Martín in Montería. Blood samples were taken from all specimens and DNA isolation carried out using salting out protocol (Miller et al., 1988), procedures took place at Animal Genetics Laboratory, Universidad de Antioquia.
Genotyping
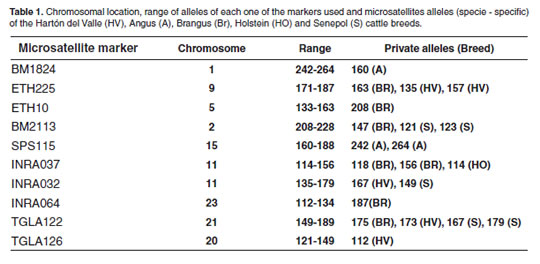
Ten microsatellite markers were used for this study. These were taken from the ISAG's recommended list for studies on genetic variability (Table 1) and were used applying multiplex systems (duplex or triplex) according to each marker's amplifi cation conditions.
Amplification conditions included approximately 50 ng of bovine genomic DNA, 10X reaction buffer (10mM Tris-HCL pH 9.0; 50mM KCl; 0.1% Tritón ® X-100), 1.5 mM of MgCl2; dNTPs; 0.125 μM of forward and reverse primers and 1 U of Taq DNA Polymerase in a total volume of 15 μL. The reaction was carried out in a T-Personal 48 thermocycler (Biometra GMBH, D-37079 Goettingen, Germany).
INRA32 and BM2113 markers were amplifi ed in duplex reaction at 58 ºC; the rest in duplex or triplex reaction at 56 ºC, under the following conditions: 94 °C for 2 min. for denaturation; next, 35 cycles at 94 °C for 45 s, 45 s for alignment, 72 °C for 45 s and final extension at 72 °C for 15 min. Amplification products were visualized in a 6% denaturing polyacrylamide gels and stained with 1% silver nitrate (Budowle et al., 1991). Genotypes designation was made by comparison of a molecular ladder.
Statistical Analysis
GDA, Genetic Data Analysis software was used for the genetic analysis (Lewis y Zaykin, 2001). This program allowed the calculation of the average number of alleles, the observed and expected heterozygosities and the Fis per breed and per locus.
Arlequín software (Schneider et al., 2000) was used for the analysis of allele frequencies (data available at request), observed and expected heterozygosities and Fis analysis.
Polymorphic Information Content (PIC) for each marker was estimated in each breed according to Botstein and White's (1980) description; the exclusion probability (EP) was calculated for both individual markers and for the group of markers per breed, according to Jamieson (1994) suggestion. These procedures were undertaken to determine whether these microsatellite markers are informative to study breed characterization and paternity tests or not.
Results
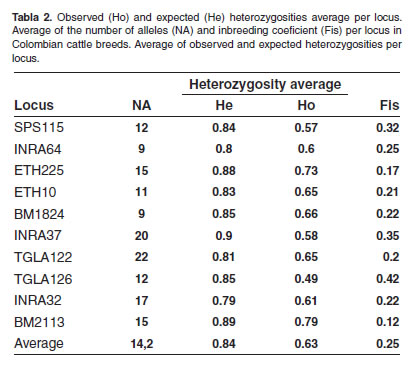
A total of 142 alleles were found in the ten analyzed loci, considering the five breed as a whole. The number of alleles per loci was between 9 (INRA64 and BM1824) and 22 (TGLA122), with an average of 14.2 (Table 2). Expected heterozygosities throughout all the breeds ranked between 0.79 (INRA32) and 0.90 (INRA37), and the average expected heterozygosity was 0.84.
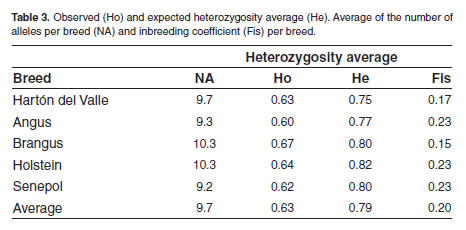
The average number of alleles per breed ranked among the breeds ranked between 0.75 in Hartón from 9.2 in Senepol breed to 10.3 in Holstein breed del Valle breed and 0.82 in Holstein breed, with an (Table 3). The expected heterozygosity range average of 0.79.
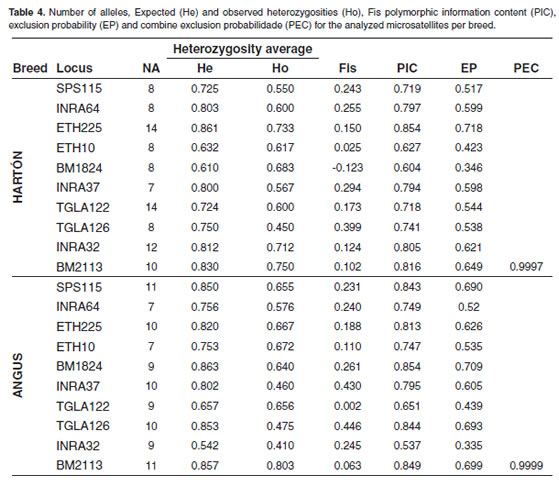
Values for the number of alleles, expected microsatellite and Angus breed showed the heterozygosity (He), observed heterozygosity lowest one in the INRA32 locus (0.410). A slight (Ho), Fis, polymorphic information content (PIC) heterozygote excess was found in the Brangus breed and exclusion probability (EP) calculated for in the INRA032 locus (-0.023) and in the Hartón each marker in each breed are shown in table 4. del Valle breed in the BM1824 locus (-0.123). PIC Senepol breed presented the lowest alleles number, varied from 0.537 for INRA32 in Senepol breed and 6 for INRA64 locus. The highest were in Brangus 0.899 for INRA37 in the same breed. The lowest for INRA37 and in Holstein for TGLA122, both combined EP was 0.9997 for Hartón del Valle with 16. Brangus breed showed the highest breed, the remaining breeds presented values higher observed heterozygosity for the BM2113 (0.845) than 0.9999.
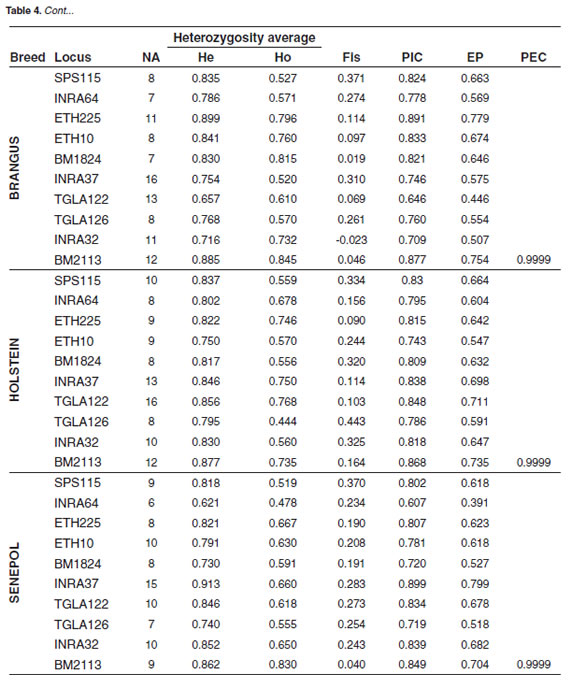
The results for EHW per breed and per locus showed that most of the loci showed disequilibrium, except for two of them (ETH10 and INRA32) in Hartón del Valle, two loci (ETH10 and TGLA122) in Angus, two loci (ETH225 y BM2113) in Holstein, two loci (INRA64 and BM2113) in Senepol and five loci (ETH10, BM1824, TGLA122, INRA32 and BM2113) in Brangus (p>0.05).
When analyzing the ten microsatellites for every breed's specific alleles, a total of three alleles were found in the TGLA122, ETH225 and INRA32 loci for Hartón del Valle; 4 alleles in the TGLA126, BM1824 and SPS115 loci for Angus; six alleles in the INRA37, BM2113, TGLA122, ETH225, INRA64 and ETH10 loci for Brangus; three alleles in BM2113, TGLA122 and INRA32 loci in Holstein and one allele in INRA37 in Senepol (Table 1). In most of the loci, the allelic frequency varied among breeds. The highest frequency was 0.20 for the 132 allele from the TGLA126 marker in Angus breed.
Discussion
In all the included breeds, all the analyzed microsatellites were polymorphic, showing between 9 (INRA64 y BM1824) and 22 alleles (TGLA122) (Table 2). In a study by Maudet et al. (2002) in native French cattle-breeds, the TGLA122 microsatellite turned out to be the most polymorphic too, with 19 alleles. Heterozygosity per locus was high and ranked between 0.79 and 0.90 (Table 2). Thus, it shows the variation degree within and among the different Colombian breeds. The difference between the average number of alleles per locus and the average number of alleles per breed might have resulted from the disparity of the analyzed samples (49 S; 60 HV, B and H, and 61A).These results show the great genetic diversity in cattle breeds in Colombia.
Genetic diversity in populations of domestic animals (cattle, buffaloes, goats, guinea pigs, etc.) has great importance from the point of view of conservation of endangered breeds. It is important to preserve genetic diversity in order for the breeds to withstand environmental changes or threats from emergent diseases, to guarantee the future supply of meat for the human population (FAO, 2007).
For most of the systems, Holstein breed showed the highest average number of alleles (Table 3), and the highest heterozygote deficit, probably due to the fact that this breed count for a huge number of specimens, but high endogamy is observed because few males are use for breeding with their daughter and granddaughters.
Senepol is a breed recently introduced to our country with a low number of specimens and even less breeding males, therefore a few numbers of alleles per locus are expected in this breed, concordant with our findings that showed a low average number of both alleles and private alleles (species - specific), in addition to the highest heterozygote deficit (Table 3). Similar results were found in Hartón del Valle breed, which is a Colombian native breed, exclusive to the Valle del Cauca region and represented by a few specimens as well, beside of this fact, this breed has been subject of conservation programs tending to preserve it because its resistance to diseases and high productivity.
Brangus crossbreed, which has been obtained from crosses between Angus and Brahman breeds, exhibited high heterozygosity values, due to the fact that has its origin from two different breeds, increasing the chance that different alleles for each locus enhance the genetic variability in Brangus breed, this might explain why Brangus breed showed the lowest heterozygotes deficit (Table 3).
An interesting outcome was the overall defi cit of heterozygotes. For all the populations, the observed heterozygosity (Ho) was lesser than expected, when analyzed per population, as well as per locus (Tables 2 and 3). This might be related with an endogamy tendency in the different herds. Artificial insemination (Maudet et al., 2002) and embryo transfer techniques also infl;uence genetically these results.
Most of the loci used in this study have been analyzed in previous studies with different breeds (Barrera et al., 2006a; Calvo et al., 2009; Hansen et al., 2002; Martínez et al., 2006; Maudet et al., 2002; Mommens et al., 1998; Radko et al., 2005). A deficit of heterozygotes only in the insemination bull population was reported by Maudet et al. (2002). In previous studies a deficit of heterozygotes was found in the Colombian native breeds Romosinuano and Hartón del Valle. (Barrera et al., 2006a), in the Colombian native breeds Romosinuano and BON (Calvo et al., 2009), A deficit of heterozygotes in Italian cattle breeds analyzed with another kind of microsatellite markers was reported by Ciampolini et al. (1995). All of these deficits have been attributed to the selection criteria for bulls chosen as sires in crossbreeding programs.
Table 4 shows the PIC values indicating the discriminatory value of each locus; values higher than 0.5 can be considered as highly discriminatory, what means that all of them allow us to value genetic diversity within and among each population. INRA37 marker for Senepol breed presented the highest PIC value and INRA32 for Angus breed presented the lowest one with 0.537.
The results of combined EP indicate that microsatellites panel must be increased for Hartón del Valle breed to a reliability minimum value of 0.9999, since its total value was 0.9997. The EP for the remaining breeds is 0.9999 and thus, this microsatellite panel could be useful to solve filiation cases (Martinez et al., 2005). Hence that the appropriate microsatellite panel for these cattle breeds parentage testing could not be adequate for other cattle breeds or vice versa; the research group considered necessary to establish a set of highly informative markers according to each breed to be evaluated.
Although some private breed alleles were found, its frequency was relatively low, except for allele 132 in Angus breed. Therefore, the possibility of using alleles for characterizing a breed would be, in principle, improbable; however, Hanotte et al. (2000) quoted by Hansen et al. (2002), has shown that it is possible to characterize a breed with relative accuracy in studies involving several cattle breeds. Steigleder et al. (2004) report the 161 allele for the ETH225 marker in the Brazilian native breed as a private allele. Further studies based on analysis involving more microsatellites or increasing the number of specimen are required in order to improve the analysis.
In conclusion, the results obtained with these microsatellites show considerable genetic diversity among the analyzed breads present in Colombia. Furthermore, microsatellites are a powerful tool for genealogy controlling. They also allow to control or organize mating systems in order to preserve their high genetic variability, and reinforce their environmental adaptation to the tropics as well as to solve filiation's cases. The combined power of exclusion for those markers exceeding 0.9999, for most of the analyzed breeds, highly suggests that this panel of markers could be theoretically considered as greatly useful for parentage verification of the analyzed cattle breeds.
Acknowledgements
We would like to thank to CODI, University of Antioquia for supporting our 2006 project Genetic Characterization of the Cattle Breeds Using Ten Microsatellite Markers. We are also grateful to Centro Internacional de Biotecnología y Reproducción Cibre, Custodiar S.A, Hacienda Sanjonhondo and Corporación Antioquia Holstein.
References
1. Barrera GP, Martínez R, Pérez JE, Polanco N y Ariza F. Evaluación de la variabilidad genética en ganado Criollo Colombiano mediante 12 marcadores microsatélites. AGRI 2006a; 38:35-45. http://www.corpoica.org.co/SitioWeb/Ofertas/ articulo.asp?id=1224 [ Links ]
2. Barrera GP, Martínez R, Torrijos R y Román F. Caracterización molecular de una población de ganado Caqueteño y su relación filogenética con razas bovinas criollas colombianas. Revista Corpoica - Ciencia y Tecnología Agropecuaria 2006b; 7:33-41. http://www.corpoica.org.co/SitioWeb/Archivos/Revista/4_ Molecular_Characterization_Caqueteo.pdf [ Links ]
3. Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment lenght polymorphism. Am J of Hum Genet 1980; 32:314-331. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1686077/pdf/ ajhg00189-0020.pdf [ Links ]
4. Budowle B, Chakraborty R, Giusti AM, Eisenberg AJ and Allen RC. Analysis of the VNTR Locus DIS80 by the PCR Followed by High-Resolution PAGE. Am J Hum Genet 1991; 48:137-144. http://www.ncbi.nlm.nih.gov/pubmed/1670750. [ Links ]
5. Calvo SJ, Martínez E, Tirado JF, Corrales JD, Montoya AE, Burgos WO, Cerón Muñoz MF, moreno M. Caracterización genética de las razas criollas BON y Romosinuano. LRRD 2009; 21. http://www.lrrd.org/lrrd21/4/calv21054.htm [ Links ]
6. Ciampolini R, Moazami-Goudarzi K, Vaiman D, Dillmann C, Mazzanti E, Foulley JL, Leveziel H, Cianci D. Individual multilocus genotypes using microsatellite polymorphisms to permit the analysis of the genetic variability within and between Italian beef cattle breeds. J Anim Sci 1995; 73:3259-3268. http://jas.fass.org/cgi/reprint/73/11/3259 [ Links ]
7. FAO. 2007. The State of the World's Animal Genetic Resources for Food and Agriculture, edited by Barbara Rischkowsky & Dafydd Pilling. Rome. [ Links ]
8. Giovambattista G, Ripoli MV, Lirón JP, Kienast ME, Villegas EE, Dulout FN, García PP. Aplicación de las técnicas de polimorfismos de DNA en la resolución de casos de abigeato, identificación individual y determinación de paternidad. Analecta Veterinaria 2001; 21: 5-11. http://www.sedici.unlp. edu.ar?id=ARG-UNLP-ART-0000000895 [ Links ]
9. Hansen C. Shrestha JN, Parker RJ, Crow GH, McAlpine PJ, et. al. Genetic diversity among Canadienne, Brown Swiss, Holstein, and Jersey cattle of Canada based on 15 bovine microsatellite markers. Genome 2002; 45:897-904. http:// article.pubs.nrc-cnrc.gc.ca/ppv/RPViewDoc?issn=08312796&volume=45&issue=5&startPage=897 [ Links ]
10. Jamieson A. The effectiveness of using codominant polymorphic allelic series for 1 checking pedigrees and 2 distinguishing full-sib pair members. Animal Genetics 1994; 25:37-44. http://www.ncbi.nlm.nih.gov/pubmed/7943982 [ Links ]
11. Kappes SM, Keele JW, Stone RT, Mcgraw RA, Sonstegard TS, Smith P.L., Lopez NL, Beattie CW. A second generation linkage map of the bovine genome. Genome Research 1997; 7:325-249. http://genome.cshlp.org/content/7/3/235.full.pdf+html [ Links ]
12. Lara MA, Contel EP, Sereno JR. Caracterización Genética de poblaciones cebuínas a través de marcadores moleculares. Arch Zootec 2005; 54:295-303. http://www. uco.es/organiza/servicios/publica/az/php/az.php?idioma_ global=0&revista=35&codigo=549 [ Links ]
13. Lewis PO, Zaykin D. (GDA) Genetic Data Analysis: Computer program for the analysis of allelic data Version 1.0 2001; http://lewis.eeb.uconn.edu/lewishome/software.html [ Links ]
14. Martínez AM, Calderón J, Camacho E, Rico C, Vega-Pla JL,Delgado JV. Caracterización genética de la raza bovina mostrenca con microsatélites. Arch Zootec 2005; 54: 357-361. http://redalyc.uaemex.mx/redalyc/pdf/495/49520738.pdf [ Links ]
15. Martínez E, Tirado JF, Cerón-Muñoz MF, Moreno M, Montoya A, Corrales JD, Calvo SJ. Caracterización genética del búfalo Murrah en Colombia usando marcadores microsatélite. Livestock Research for Rural Development 2009; 21. http://www.lrrd.org/lrrd21/1/mart21014.htm [ Links ]
16. Martínez R, Barrera G, Sastre H. Variabilidad y estado genético de siete subpoblaciones de la raza criolla colombiana Casanareño. Revista Corpoica - Ciencia y Tecnología Agropecuaria 2006; 7: 5-11 http://www.corpoica.org.co/SitioWeb/Revistas/verarticulo.asp?id_contenido=209 [ Links ]
17. Maudet C, Luikart G, Taberlet P. Genetic diversity and assignment tests among seven French cattle breeds based on microsatellite DNA analysis. J Anim Sci 2002; 80:942-950. http://jas.fass.org/cgi/reprint/80/4/942.pdf [ Links ]
18. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cell. Nucleic Acids Research 1988; 16:1215. http://www.ncbi.nlm. nih.gov/pmc/articles/PMC334765/pdf/nar00145-0424.pdf [ Links ]
19. Mommens G; Van A; Peelman LJ. Effectiveness of bovine microsatellites in resolving paternity cases in American bison, Bison bison. Animal Genetics 1998; 29:12-18. http:// www3.interscience.wiley.com/cgi-bin/fulltext/119119507/ PDFSTART [ Links ]
20. Moreno F, Bedoya G, Derr J, Carvajal L, Bermúdez N, Zuluaga N, Ossa J, Berdugo J, Estrada L, Barrera J, Scott D, Linares A. Diversidad genética y relaciones fi logenéticas del ganado criollo colombiano. Revista Corpoica 2001; 3:17-23 http://www.corpoica.org.co/SitioWeb/Archivos/Revista/3_ Diversidadgenticayrelacion.PDF [ Links ]
21. Peelman LJ, Mortiaux F, Van A, Dansercoer A, Mommens G, Coopman F, Bouquet Y, Burny A, RenavIle R, Portetelle D. Evaluation of the genetic variability of 23 bovine microsatellite markers in four Belgian cattle breeds. Animal Genetics 1998; 29:161-16. http://cat.inist.fr/?aModele=afficheN&cpsi dt=2342763 [ Links ]
22. Radko A, Zyga A, Zabek T, Slota E. Genetic variability among Polish Red, Hereford and Holstein-Friesian cattle raised in Poland based on analysis of microsatellite DNA sequences. Short communication. J Appl Genet 2005; 46:89-91. http://jag.igr.poznan.pl/2005-Volume-46/1/pdf/2005_ Volume_46_1-89-91.pdf [ Links ]
23. Sanz A, Uffo O, Miranda I, Martínez S. Empleo de los microsatélites para determinar paternidad en Bovinos Cubanos. Rev Salud Anim 2002; 24:166-169. http://www. censa.edu.cu/index2.php?option=com_docman&task=doc_ view&gid=574&Itemid=105 [ Links ]
24. Steigleder CS, Almeida EA, Weimer TA. Genetic diversity of a Brazilian creole cattle based on fourteen microsatellite loci. Arch Zootec 2004; 53:3-11. http://redalyc.uaemex.mx/redalyc/ pdf/495/49520101.pdf [ Links ]
25. Zamorano MJ, Género ER, Rodero A, Vega-Pla JL, Rumiano FJ. Caracterización genética de ganado bovino criollo argentino utilizando microsatélites. Arch Zootec 1998a; 47: 273-277. http://www.uco.es/organiza/servicios/publica/az/ articulos/1998/178-179/pdf/zamoran3.pdf [ Links ]
26. Zamorano MJ, Ruiter J, Rodero A, Vega-Pla JL. Análisis genético de marcadores microsatélites en dos poblaciones de la raza bovina berrenda en negro. Arch Zootec 1998b; 47:195-200. http://www.uco.es/organiza/servicios/publica/az/ articulos/1998/178-179/pdf/zamoran2.pdf [ Links ]