Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Acta Biológica Colombiana
Print version ISSN 0120-548X
Acta biol.Colomb. vol.17 no.1 Bogotá Jan./Apr. 2012
A NEW WORLD MONKEY MICROSATELLITE (AP74) HIGHLY CONSERVED IN PRIMATES
AP74, un microsatélite de Monos del Nuevo Mundo altamente conservado en Primates
LUCIANA INÉS OKLANDER1, 2, 4*, Ph. D.; ELIANA RUTH STEINBERG3,4,*, Ph. D.; MARTA DOLORES MUDRY3, 4,*, Ph. D.
1 IBS (Instituto de Biología Subtropical) Facultad de Ciencias Forestales, Universidad Nacional de Misiones. Puerto Iguazú, Misiones. Argentina. .
2 Servicio de Huellas Digitales Genéticas, Facultad de Farmacia y Bioquímica, Universidad de Buenos Aires, Argentina.
3 Grupo de Investigación en Biología Evolutiva, Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires, Argentina.
4 CONICET
* These authors contributed equally to this work.
Full Corresponding Author address: Luciana Inés Oklander, IBS (Instituto de Biología Subtropical) Facultad de Ciencias Forestales, Universidad Nacional de Misiones. Andresito 21, Puerto Iguazú, Misiones. Argentina. Tel/Fax: 54 11 4313 3574; lulaok@gmail.com
Presentado 25 de febrero de 2011, aceptado 28 de octubre de 2011, correcciones 4 de noviembre de 2011.
ABSTRACT
Given their great variability, microsatellites or STRs became the most commonly used genetic markers over the last 15 years. The analysis of these markers requires minimum quantities of DNA, allowing the use of non invasive samples, such as feces or hair. We amplified the microsatellite Ap74 in blood and hair samples in order to analyze the levels of genomic conservation among a wide range of primates including: Lemur catta, Alouatta caraya, Ateles belzebuth, Ateles chamek, Pan troglodytes, Papio sp., and Homo sapiens. In all cases we obtained amplification products that exhibited similar size both in monkeys and human (oscillating between 126 and 176 bp), except in the lemur where the detected fragment presented a size of approximately 1000 bp. The analysis of the nucleotide sequences permitted the evaluation of the molecular modifications experienced during the evolutionary process in primates.
Key words: Primates, microsatellite markers, PCR amplification, non-invasive sampling, genomic conservation, genetic variability.
RESUMEN
Dado su alta variabilidad, los microsatélites o STR se convirtieron en los marcadores genéticos más ampliamente utilizados en los últimos 15 años. El análisis de estos marcadores requiere una mínima cantidad de ADN, permitiendo el uso de muestras no invasivas, tales como pelos o heces. Con el objetivo de analizar niveles de conservación genómica, amplificamos el microsatélite Ap74 en muestras de pelo y sangre de un amplio rango de primates incluyendo: Lemur catta, Alouatta caraya, Ateles belzebuth, Ateles chamek, Pan troglodytes, Papio sp., y Homo sapiens. En todos los casos obtuvimos productos de amplificación que exhibieron un tamaño similar (oscilando entre 126 y 176 pb), con excepción del lémur, donde el fragmento detectado presentó un tamaño de aproximadamente 1000 pb. El análisis de las secuencias nucleotídicas nos permitió evaluar las modificaciones moleculares ocurridas durante el proceso evolutivo en primates.
Palabras clave: Primates, marcadores microsatélites, amplificación por PCR, muestreo no-invasivo, conservación genómica, variabilidad genética.
INTRODUCTION
Molecular techniques have enlarged the underlying knowledge about the evolutionary process. Microsatellites, also called short tandem repeats (STRPs), are useful genetic markers for the study of the demographic structure and the phylogenetic history of the populations. These markers are scattered throughout the vertebrate’s genome and are highly polymorphic, varying in the number of motif`s repetition. As they generally seem to be free of selective constraints, it is evident that this extensive degree of genetic variability requires a high underlying mutation rate (Schlötterer, 2000; Ellegren, 2004). The analysis of these markers is based on the polymerase chain reaction (PCR) and requires minimum quantities of DNA, allowing the use of non invasive samples, such as feces or hair (Frantzen et al., 1998; Taberlet et al., 1999; Oklander et al., 2004). Since their flanking sequences are highly conserved, the employment of heterologous primers allows the detection of diverse sequences conserved across related species.
Since the first phylogenetic analysis employing microsatellites as a new tool was published fifteen years ago, the information on the evolution and molecular dynamics of these markers in different species is constantly increasing (Rogers et al., 1995; Menottiraymond and O Brien, 1995; Clisson et al., 2000; Neff and Gross, 2001; Lathuilliere et al., 2001; Oklander et al., 2006; Ruiz García et al., 2005; Ruiz García et al., 2006; Ruiz García et al., 2007).
Microsatellites isolated for humans allowed the amplification of sequences in several species of nonhuman primates including apes, baboons, macaques, and few plathyrrine monkeys (Blanquer-Maumont and Crouau-Roy, 1995; Garza et al., 1995; Coote and Bruford, 1996; Kayser et al., 1996; Ellsworth and Hoelzer, 1998; Rogers et al., 2000; Goossens et al., 2000; Nair et al., 2000; Smith et al., 2000). Conservation of the sequences among groups of taxa is limited, since numerous substitution events and/or insertion/deletion (that don’t always happen at random) do occur, and they can modify the molecular structure from regions near to the microsatellites (Clisson et al., 2000; Ruiz Garcia, 2005). The goal of this study was to evaluate the level of conservation of one microsatellite, isolated for a New World Monkey, among many primate species.
MATERIALS AND METHODS
DNA was extracted from samples of two origins: a) blood samples conserved in FTA (Whatman) of the species Alouatta caraya (ACA, of a wild specimen from Isla Brasilera, Corrientes, Argentina, within the natural distribution of the species), Ateles belzebuth (ABE, in captivity at the Zoo of Buenos Aires, Argentina), Ateles chamek (ACH, in captivity at the Zoo of Buenos Aires, Argentina) and Homo sapiens (HSA, control sample taken from a GIBE member), following the protocol provided by the supplier and b) hair, from specimens in captivity at the Zoo of La Plata (Buenos Aires, Argentina), of Lemur catta (LCA), Pan troglodytes (PTR) and Papio sp. (PSP), using standard phenol-chloroform extraction followed by purification and concentration of the extracted DNA from hair by means of columns Microcon YM-100 (Millipore).
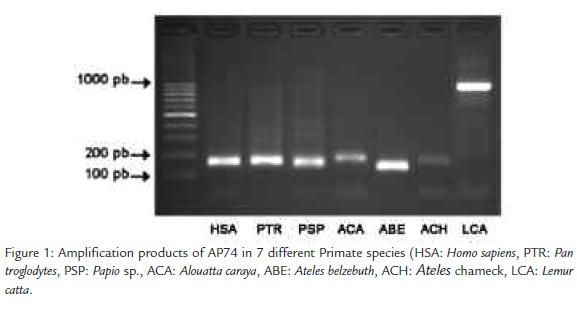
We amplified one microsatellite isolated from Alouatta palliata (AP74). Primers used were: (5’-TGCACCTCATCTCTTTCTCTG-3’) and (5’-CATCTTTGTTTTCC TCATAGC - 3’; Ellsworth and Hoelzer, 1998). DNA amplifications were performed in a total volume of 25 µl (20 mM Tris-HCl, 50 mM KCl, and 1.5 mM MgCl2, 0.2 mM each dNTP, 1 U Taq DNA Polymerase (Tandil), 4 pmol of each primer and 1 sheet of FTA or l ml of the extracted DNA). PCR reaction consisted on 35 cycles of 1 min at 95 °C, 1 min to 52 °C, and 1 min 30 s at 72 °C. PCR products were run in 2% agarose gels, dyed with ethidium bromide and sequenced by means of the method Big dye terminator system (Applied Biosystems). Sequences were analyzed in an automatic secuenciator (ABI Prism 310). The DNA sequences obtained were aligned using Clustal W and the Neighbourjoining analysis was made using MEGA version 3.0 (Kumar et al., 2004).
RESULTS
We obtained amplification products of similar size (between 126 and 176 bp)in all the analyzed primates, with the exception of Lemur catta, which presented a size of approximately 1000 pb (Figura 1). We successfully sequenced the fragments obtained for all the analyzed species.
The analysis of the sequences among these species showed a high conservation of the flanking regions, in contrast with the great variation found in the number of repetitions of the microsatellite (CA; Figura 2). Among New and Old World Primates, inserts, transitions and transversions were observed in the later region of the microsatellite. Also, punctual substitutions were detected in species from the same genus (Ateles) and in species phylogenetically closer as Homo sapiens and Pan troglodytes that were absent in the rest of the primates DNA studied (Figura 2). Lemur catta showed only limited alignment with the other DNA sequence in the 3’ extreme. The DNA sequence detected for Homo sapiens was searched in Gene Bank database and was located in chromosome 4 (clone RP11-752D24), among bases 169871 and 170030. Bold: Primer sequences, Gray: repetitive area of the microsatellite, the points are conserved bases across primate species and the dashes are gaps.
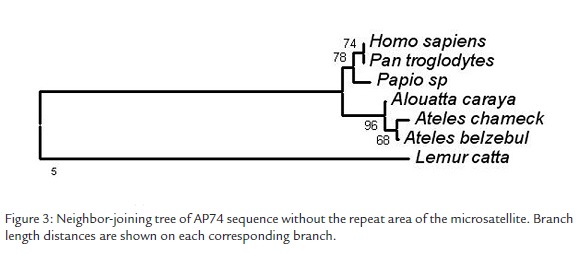
With the differences found among species we made a Neighbor-joining analysis based on TrN+G genetic distances using MEGA version 3.0 (Kumar et al., 2004). The DNA sequence used as reference was the one published for Alouatta paliatta (NCBI U36400) that possesses a great number of undetermined bases. Given this fact, this DNA sequence was not included in the neighbor joining analysis. A consensus parsimony cladogram (PAUP*, Swofford, 2002; not shown) was fully concordant with the NJ phenogram. The robustness of the nodes was determined by means of a bootstrap analysis of 1000 replicates. The support of the node among monkeys of the New and Old World was of 96%, between Homo sapiens and Pan troglodytes of 62% and among species of the same genus of 66% (Figura 3). The differences found in the repeat area of the microsatellite were not considered informative since it is ignored whether the number of repetitions is species-specific or it possesses intraspecific variation.
DISCUSSION
Many are the mechanisms and strategies that superior organisms present as frequent and characteristic of the speciogenic process (for a review see Sobel et al., 2009 and references therein). It cannot be said that there is a unique mechanism. Initialy, in order to understand the dynamic of speciation at the population level, molecular genetics appeared to be a more informative tool than classical genetics (morphological data through chromosomal rearrangements).
In vertebrates, and particularly in primates, phylogenetic and phylogeographic studies were initiated (both in Old and New World Primates) recurring to mitochondrial DNA analysis, considering that these genetic markers could be studied at the population level in order to implement the right policies for the conservation of the current species (Collins et al., 2000; Ascunce et al., 2003a; Ascunce et al., 2007). In New World Primates, COII cytochrome analysis constituted an important tool to perform phylogenetic inferences (Ascunce et al., 2003b; Ascunce et al., 2003c), as well as cytochrome b was informative for a better understanding of the speciogenic process associated to geographic distribution in several species (Lavergne et al., 2003).
However, the appearance of microsatellite studies allowed to enlarge and deepen the knowledge about the changes that took place at genomic level and that are related to species` evolutionary processes. In Primates, species-diagnostic microsatellites loci were used to determine hybrid status in two species of neotropical monkeys from an area of sympatry in Mexico where they have not achieved complete isolation (Cortes Ortiz et al., 2007). Another interesting case was referred employing DNA microsatellites analyzing gene diversity and bottleneck events in different neotropical monkey populations with conservation purposes (Ruiz Garcia et al., 2006; Ruiz Garcia et al., 2007).
In this contribution, the study of sequences flanked by conserved ones allowed us to evaluate molecular modifications during the evolutionary process in primates. Since the fragments obtained for species extremely distanced as Homo sapiens and Alouatta caraya possess an extremely similar size it was necessary to analyze the composition of sequences in order to identify the substitution insertion or deletion events. The results obtained for the sequence analysis confirmed the evolutionary conservation of this locus in the Order Primates.
Generally crossed amplifications between species is successful at the Genus or Family level and less frequently at the Order level (Clisson et al., 2000; Ruiz Garcia et al., 2006; Ruiz Garcia et al., 2007). Such is the case of Coote and Bruford, 1996, who could not amplify microsatellites isolated from humans in Prosimian and New World primates. In the present contribution, the marker AP74 allowed the amplification of sequences in several species of the Primates Order.
The microsatellite AP74 was located to chromosome 4, but it was not possible to perfom an assignment to its correspondant chromosome band (Genbank). Considering that in genomic conservation analysis regions and bands of the human karyotype have been establish as highly conserved among New World and Old World Primates (Weinberg et al., 2005; Stanyon et al., 2008 and references therein) we can suggest that the sequences referred in this contribution could be located on one of the following chromosomes of each analyzed species: Pan troglodytes: PTR3 (Jauch et al., 1992); Papio sp.: PSP 6 (Rogers et al., 2000); A. caraya: ACA4q, ACA 9 and ACA22qter (de Oliveira et al., 2002); Ateles chamek: ACH2q12-q14, ACH8p and ACH15 (Ruiz-Herrera et al., 2005); Ateles belzebul: ABE2q12-q14, ABE9p and ABE15 (García et al.,, 2002); Lemur catta: LCA5pprox, LCA4p, LCA19ter, LCA23, LCA26 (Warter et al., 2005).
Previous studies have shown that alterations in the base composition of the repetitions is an important component of the variation between individuals and species (Garza et al., 1995; Crouau-Roy et al., 1996). This study was carried out with only one sample by species, therefore the obtained number of repetitions of the reason CA is not necessarily characteristic of each species. Further studies with more individuals are needed to determine if indeed what is reported in the present study ocurred at population level. In this manner we will be evaluating the levels of variation within and between populations in order to clarify the possible genomic conservation as well as the phylogenetic reconstruction in these primate taxa.
Our study contributed to the permanent evaluation and correction of the phylogenetic history of the Primate species. Furthermore, new studies involving microsatellites sequence analysis across related species will allow further implementation of these markers in evolutionary studies.
ACKNOWLEDGEMENTS
All research reported in this manuscript has met the appropriate national and institutional guidelines for the legal acquisition and use of laboratory animals and authorized study of wild animals. The authors also adhered to the Guide for Care and Use of Experimental Animals as promulgated by the Canadian Council of Animal Care and to the American Society of Primatologists (ASP) Principles for the Ethical Treatment of Non Human Primates. All authors approved the submission of the manuscript and have no conflicts of interest regarding its publication. Our gratitude to Dr. Daniel Corach for allowing us to perform the studies in his laboratory. We thank the authorities of the institutions and the veterinarians of the Zoo of La Plata and the Zoo of Buenos Aires for their assistance in the sampling of the animals.
This research was supported by grants to MDM from the Argentinean National Council for Scientific and Technological Research (CONICET PIP 5012) and Buenos Aires University (UBACyT X031).
REFERENCES
ASCUNCE MS, CORTES ORTIZ L, MUDRY MD. The mitochondrial control region of the black howler monkey, Alouatta caraya (Primates, Platyrrhini), and the development of new primers. Mol Ecol Notes. 2003a;3:372-375. [ Links ]
ASCUNCE MS, HASSON E, MUDRY MD. COII: a useful tool for inferring phylogenetic relationships among New World Monkeys (Primates, Platyrrhini). Zool Scripta. 2003b;32(5):397-406. [ Links ]
ASCUNCE MS, OKLANDER LI, MUDRY MD. Amplification of Mitochondrial COII Gene from DNA Extracted from Hair Samples in Some Species of New World Monkeys. Fol Primatol. 2003c;74:165-167. [ Links ]
ASCUNCE MS, HASSON E, MULLIGAN CJ, MUDRY MD. Mitochondrial sequence diversity of the southernmost extant New World Monkey, Alouatta caraya. Mol Phylogenet Evol. 2007;43:202-215. [ Links ]
BLANQUER-MAUMONT A, CROUAU-ROY B. Polymorphism, monomorphism, and sequences in conserved microsatellites in primate species. J Mol Evol. 1995;41(4):492-497. [ Links ]
CLISSON I, LATHUILLIERE M, CROUAU-ROY B. Conservation and evolution of microsatellite loci in primate taxa. Am J Primatol. 2000;50:205-214. [ Links ]
COLLINS AC, DUBACH JM. Phylogenetic relationships of Spider Monkeys (Ateles) based on Mitochondrial DNA variation. Am J Primatol. 2000;21(3):981-420. [ Links ]
COOTE T, BRUFORD MW. Human microsatellites applicable for analysis of genetic variation in apes and Old World Monkeys. J Hered. 1996;87:406-410. [ Links ]
CORTES-ORTIZ L, DUDA TF JR, CANALES-ESPINOSA D, GARCIA-ORDUNA F, RODRIGUEZ-LUNA E, BERMINGHAM E. Hybridization in Large-Bodied New World Primates. Genetics. 2007;176:2421-2425. [ Links ]
CROUAU-ROY B, SERVICE S, SLATKIN M, FREIMER NB. A fine scale comparison of the human and chimpanzee genomes: linkage, linkage desequilibrium and sequence analysis. Hum Mol Genet. 1996;5:1131-1137. [ Links ]
DE OLIVEIRA EHC, NEUSSER M, FIGUEIREDO WB, NAGAMACHI C, PIECZARKA JC, SBALQUEIRO IJ, et al. The phylogeny of howler monkeys (Alouatta, Platyrrhini): reconstruction by multicolor or cross-species chromosome painting. Chrom Res. 2002;10:669-683. [ Links ]
ELLEGREN H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435-445. [ Links ]
ELLSWORTH JA, HOELZER GA. Characterization of microsatellite loci in a New World Pprimate, the mantled howler monkey (Alouatta palliata). Mol Ecol.1998;7(5):657-658. [ Links ]
FRANTZEN MAJ, SILK JB, FERGUSON JWH, WAINE RK, KOHN MH. Empirical evaluation of preservation methods for faecal DNA. Mol Ecol. 1998;7:1423-1428. [ Links ]
GARCÍA F, RUIZ-HERRERA A, EGOZCUE J, PONSA M, GARCÍA M. Chromosomal homologies between Cebus and Ateles (Primates) based on ZOO-FISH and G-banding comparisons. Am J Primatol. 2002;57(4):177-188. [ Links ]
GARZA JC, SLATKIN M, FREIMER NB. Microsatellite allele frequencies in humans and chimpanzees with implications for constrain in allele size. Mol Biol Evol. 1995;12: 594-603. [ Links ]
GOOSSENS B, LATOUR S, VIDAL C, JAMART A, ANCRENAZ M, BRUFORD M. Twenty new microsatellite loci for use with hair and faecal samples in the chimpanzee (Pan troglodytes troglodytes). Folia Primatol. 2000;71:177-180. [ Links ]
JAUCH A, WIENBERG J, STANYON R. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc Nat Acad Sci U S A. 1992;89:8611-8615. [ Links ]
KAYSER M, RITTER H, BERCOVITCH F, MRUG M, ROEWER L, NURNBERG P. Identification of highly polymorphic microsatellites in the rhesus macaque Macaca mulatta by cross-species amplification. Mol Ecol. 1996;5:157-159. [ Links ]
KUMAR S, TAMURA K, NEI M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5(2):150-163. [ Links ]
LATHUILLIERE M, MÉNARDA N, CROUAU-ROYB B. Sequence conservation of nine Barbary Macaque (Macaca sylvanus) Microsatellite Loci: implication of specific primers for genotyping. Folia Primatol. 2001;72:85-88. [ Links ]
LAVERGNE A, CATZEFLIS F, LACOTE S, BARNAUD A, BORDIER M, MERCEREAUPUIJALON O, et al. Genetic analysis of the Saimiri breeding colony of the Pasteur Institute (French Guiana): Development of a molecular typing method using a combination of nuclear and mitochondrial DNA markers. J Med Primatol. 2003;32;330-340. [ Links ]
MENOTTI-RAYMOND MA, O BRIEN SJ. Evolutionary conservation of ten Microsatellite loci in fourn species of Felidae. J Hered. 1995;86:319-322. [ Links ]
NAIR S, HA J, ROGERS J. Nineteen new microsatellite DNA polymorphisms in pigtailed macaques (Macaca nemestrina). Primates. 2000;41:343-350. [ Links ]
NEFF BD, GROSS MR. Microsatellite evolution in vertebrates: inference from ac dinucleotide repeats. Evolution. 2001;55(9):171. [ Links ]
OKLANDER LI, MARINO M, ZUNINO GE, CORACH D. Preservation and extraction of DNA from feces in Howler Monkeys (Alouatta caraya). Neotrop Primates. 2004;12(2):59-63. [ Links ]
OKLANDER LI, ZUNINO GE, DI FIORE A, CORACH D. Isolation, characterization and evaluation of 11 autosomal STRs suitable for population studies in black and gold howler monkeys Alouatta caraya. Mol Ecol Notes. 2006;7:117-120. [ Links ]
ROGERS J, MAHANEY MC, WITTE SM, NAIR S, NEWMAN D, WEDEL S, et al. A genetic linkage map of the baboon (Papio hamadryas) genome based on human microsatellite polymorphisms. Genomics. 2000;67:237-247. [ Links ]
ROGERS J, WITTE SM, SLIFER MA. Five new Microsatellite DNA polymorphisms in squirrel monkey (Saimiri boliviensis). Am J Primatol. 1995;36:1517-1733. [ Links ]
RUIZ-GARCÍA M. The use of several microsatellite loci applied to 13 Neotropical Primates revealed a strong recent bottleneck event in the woolly monkey (Lagothrix lagotricha) in Colombia. Primate Report. 2005;71:27-55. [ Links ]
RUIZ-GARCÍA M, PARRA A, ROMERO-ALEAN N, ESCOBAR-ARMEL P, SHOSTELL J. Genetic Characterization and phylogenetic relationships between the Ateles species (Atelidae, Primates) by means of DNA microsatellite markers and craniometric data. Primate Report. 2006;73:3-47. [ Links ]
RUIZ-GARCÍA M, ESCOBAR-ARMEL P, ALVAREZ D, MUDRY MD, ASCUNCE M, GUTIERREZ-ESPELETA G. Genetic variability in four Alouatta species measured by means of nine DNA microsatellite markers: Genetic structure and recent bottlenecks. Folia Primatol. 2007;78:73-87. [ Links ]
RUIZ-HERRERA A, GARCÍA F, MORA L, EGOZCUE J, PONSÁ M, GARCÍA M. Evolutionary conserved chromosomal segments in the human karyotype are bounded by unstable chromosome bands. Cytogenet Genome Res. 2005;108:161-174. [ Links ]
SCHLÖTTERER C. Evolutionary dynamics of microsatellite DNA. Chromosoma. 2000;109:365-371. [ Links ]
SMITH DG, KANTHASWAMY S, VIRAY J, CODY L. Additional highly polymorphic microsatellite (STR) loci forestimating kinship in rhesus macaques (Macaca mulatta). Am J Primatol. 2000;50(1):1-7. [ Links ]
SOBEL JM, CHEN GF, WATT LR, SCHEMSKE W. The biology of speciation. Evolution .2009;64(2):295-315. [ Links ]
STANYON R, ROCCHI M, CAPOZZI O, ROBERTO R, MISCEO D, VENTURA M, et al. Primate chromosome evolution: Ancestral karyotypes, marker order and neocentromeres. Chrom Res. 2008;16(1):17-39. [ Links ]
SWOFFORD DL. PAUP*. Phylogenetic analysis using parsimony and other methods version 4. Sunderland, Massachusetts, Sinauer; 2002. [ Links ]
TABERLET P, WAITS LP, LUIKART G. Noninvasive genetic sampling: look before you leap. Trends Ecol Evol. 1999;14:323-327. [ Links ]
WARTER S, HAUWY M, DUTRILLAUX B, RUMPLER Y. Application of molecular cytogenetics for chromosomal evolution of the Lemuriformes (Prosimians). Cytogenet Genome Res. 2005;108:197-203. [ Links ]
WEINBERG J. Fluorescent in situ hybridization to chromosomes as a tool to understand human and primate genome evolution. Cytogenet Genome Res. 2005;108: 139-160. [ Links ]