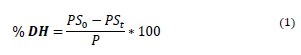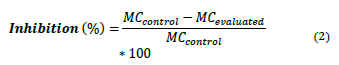1. Introduction
Microalgae are photosynthetic microorganisms [1], producers of a wide variety of metabolites of interest such as lipids, carbohydrates, proteins, pigments, vitamins, among others [2-6], playing an important role in diverse applications as cosmetic, pharmaceutical industries, biofuels, biofertilizers, wastewater treatment [7].
The proteins, protein hydrolysates and peptides from an important variety of strains of microalgae have received special interest for their antioxidant, anticancer, antihypertensive and antimicrobial activity, etc. [1-4]
The mechanism of action of conventional antibiotics coupled with the excessive and improper use of these compounds has led to the increase of resistant and multiresistant microorganisms, hindering the treatment and control of infectious diseases. For this reason, a promising solution to this problem is to find compounds with different mechanisms of action such as antimicrobial peptides (AMP). AMPs are found in various sources, such as microorganisms, plants, vertebrates or invertebrates [8]. These compounds are part of the innate immune system of a large variety of individuals, playing a role against the attack of invading organisms [9]. Their broad spectrum of action gives them an important effect against bacteria, fungi, viruses, and parasites. Their structural diversity allows them to act against several biological targets such as cell membranes, proteins, nucleic acids, and enzymes [8-10].
Specifically, the potential of different peptides and protein hydrolysates of microalgal origin has been reported by various authors as possible therapeutic agents from an unconventional source, due to its antimicrobial [11], antiviral [12], immunoregulatory function [13,14], with a wide range of applications.
In this context, this work focuses on obtaining a hydrolyzed protein extract from the microalgae Nannochloropsis sp., and to determine its antimicrobial activity against Staphylococcus aureus, Escherichia coli, and Candida albicans.
2. Materials y methods
2.1. Biomass obtaining
The concentrated biomass of Nannochloropsis sp. was obtained from a culture carried out in 250L photobioreactors, under continuous light conditions with white fluorescent lamps (135 μmolm2s-1) and a temperature of 23 ± 1°C. The culture medium used was agricultural fertilizer (NPK 13:40:13). After 11 days of cultivation, the biomass was washed between 3 and 5 times to remove the excess of salts and subsequently dried in a WTC BINDER oven at 50°C. It was manually macerated in a mortar to obtain a fine powder and stored in a dry place until its use.
2.2. Extraction and protein quantification
60 g of dry biomass was suspended in 600mL of 50 mM phosphate buffer at pH 7, the mixture was reserved at 4°C for 15 minutes. Subsequently, a mechanical process corresponding to homogenization was performed in an ULTRA TURRAX IKA T25 DIGITAL unit, at 6000 rpm, during 20 cycles of 5-minute on and 2-minute intervals. During homogenization, the mixture was kept in an ice bath, to prevent its heating. The extract was centrifuged at 10,000 rpm for 10 minutes and vacuum filtered with a cellulose membrane filter of 0.2 μm pore diameter. Finally, the supernatant was subjected to dialysis for 24 h, using a membrane with a pore size of 3.5 kDa of Spectra/Por (Spectrum laboratories) obtaining the crude extract.
The crude protein present in the biomass was analyzed by the percentage of total nitrogen by the Kjendhal method, using 5.2 as a conversion Nitrogen-to-protein factor [15].
The soluble protein was determined by the Lowry method [16], using a calibration curve with BSA (bovine serum albumin) between 0.1 and 1 mg/mL. The sample was analyzed in a UV-Vis Spectronic Genesys 2PC equipment, at a wavelength of 750 nm.
2.3. Enzymatic hydrolysis
The protein extract was hydrolyzed with a mixture of Papain (PROENZIMAS) and Pancreatin (Abbott Laboratories), at 44 ° C and pH 7, for 6 hours, at an enzyme-substrate ratio of 10 U/g of protein for each enzyme [17]. Subsequently, the hydrolyzed extract was passed through a 3 kDa AMICON filter.
The degree of hydrolysis (DH) was quantified by equation 1, which is presented below:
% DH: Degree of hydrolysis (%).
PS0: Amount of soluble protein in 6.25%(w/v) TCA (Trichloroacetic acid), before the addition of enzyme.
PSt: Amount of soluble protein in 6.25%(w/v) TCA after the addition of enzyme, in a t time.
P: Amount of initial protein.
2.4. Polyacrylamide gel electrophoresis (SDS-PAGE)
The samples were concentrated by adding to 10 mL of them, 10 mL of 20% (w/v) TCA [18], left at 4°C overnight, centrifuged at 15,000 rpm for 15 minutes at 4°C and the obtained pellet was washed with 300 μL of cold acetone. They were centrifuged again at 15,000 rpm for 15 minutes at 4°C and the supernatant was removed. The resulting pellet was suspended in 60μL of solubilization Buffer (100 mM Tris, 100 mM EDTA and 8M Urea) pH 8.5.
To the previous mixture was added 20μL of a solution composed of 10μL of 2-mercaptoethanol and 90μL of coloring solution (SDS, Coomasie Blue R-250, glycerol) and left in incubation for 5 minutes at 80°C. Then, 15μL were seeded in the SDS Tris-tricine gel at 14% concentration and the electrophoretic run was carried out in a Bio-Rad Mini-PROTEAN® equipment at 150 V, for 1 h and 15 minutes.
The resulting gel was stained in a Coomassie Blue R-250 solution for 12 hours under constant agitation. The molecular marker used was the Blue Prestained Protein Standard from Biolabs® Inc., with a range between 11-190 kDa.
2.5. Fourier transform infrared spectroscopy - FTIR
The extracts were analyzed in a Bruker Tensor II FT-IR with diamond ATR (Bruker Optics, Germany). The equipment consists of a photovoltaic MCT detector cooled with liquid nitrogen. The wavenumber range was from 4,000 to 700 cm-1 with a resolution of 4 cm-1. The curve fitting was analyzed using the OPUS Software (Bruker Optics, Germany). The baseline of the spectra was corrected and analyzed in each selected region.
2.6. Antimicrobial activity
The broth dilution method [19,20] was implemented, using the following microorganisms: Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 25923) and Candida albicans (ATCC 90028). The fraction less than 3 kDa of the hydrolyzed extract was passed through a 0.2 μm cellulose membrane filter to eliminate possible contaminants.
The strains were activated in broth, TSB for bacteria and Sabouraud for yeasts, at a temperature of 36°C for 24 h. The inoculum was adjusted with a 0.5 McFarland pattern.
In tubes of 5 mL capacity, were added 3 different concentrations of hydrolyzed extract, 33.35% (v/v), 20% (v/v) and 10% (v/v), 1233 μL of broth (TSB or Sabouraud), and finally, 100 μL of the inoculum, respectively. The tests were carried out in triplicate. Additionally, a feasibility control was made for each microorganism, which did not contain the extract. The samples were placed in incubation for 24 h at 36°C. Serial dilutions were made in triplicate in 0.9% (w/v) saline solution from each sample and 10 μL of each one was seeded in TSA agar (bacteria) or PDA (yeast) according to the micro drop technique [21]. The samples were left in incubation at 36°C, 24 h for bacteria and 48 h for yeast. Finally, it was made a recount of the colonies present in each sowed dilution.
The percentage of inhibition of the strains was calculated [22], according to equation 2:
MC control: Number of colonies forming units per milliliter of the control treatment. (CFU/mL)
MC evaluated: Number of colonies forming units per milliliter of the evaluated treatment. (CFU/mL)
3. Results and discussion
3.1. Cellular lysis and protein release
Nannochloropsis is a genus widely used at the industrial level because of their high nutritional quality and additionally being a source of value-added compounds such as pigments, polyunsaturated fatty acids [23] and, of course, proteins and peptides with biofunctionality [24] applicable to the field of animal and human health. Even though the quantity and quality of proteins is not a problem, the method of extracting them is the principal complication in the release of these compounds, due to the presence of the cell wall composed of two layers, of which the cellulose is one of the main components [25].
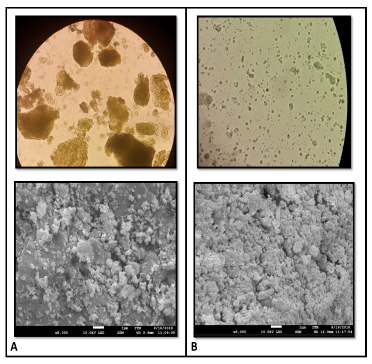
Figure 1 shows the beginning and end of mechanical cell disruption treatment. Samples from the upper side of the figure were visualized with an Olympus CX21 microscope with a 40X objective and those from the bottom, with a scanning electron microscope (SEM) at 8000X.
Source: The Authors.
Figure 1 Cells of Nannochloropsis sp. before and after homogenization treatment. A) Before treatment. B) Cycle 20.
Because of the previous drying process, the biomass before starting the treatment shows agglomerations of cells, which obstruct the lysis of the cell wall. In cycle 20 of the treatment, these accumulations are eliminated, allowing the cell to be more vulnerable to mechanical stresses and improve the probabilities of disruption. The increase in surface area and the heterogeneity in cell sizes are evidence of the effect of the mechanical action of lysis on the cell surface.
The results show that the initial crude protein of the biomass was 8.84%. The mechanical extraction process achieved a percentage of cell lysis of 60.1 ± 3.3% and a soluble protein concentration in the extract of 0.88 ± 0.03%, getting an extraction yield of 9.9 %.
The extraction yield found in this study was very similar to the one got by Zhang et al. [26], who obtained a yield of 12.1% when submitting 1 g of Chlorella pyrenoidosa biomass to a homogenization system at 8,000 rpm for 10 minutes and 2-minute intervals. However, it should be noted that the amount of biomass used in this study was greater. On the other hand, some authors have reported higher extraction yields when implementing other mechanical methods such as high- pressure homogenization and grinding. Safi et al. [15] reported a 95% percent lysis and 50% (w/w) protein release in Nannochloropsis gaditana cells, under a high-pressure homogenizer; while Pan et al. [27] achieved a percentage of cell lysis superior to 98% on Nannochloropsis sp. using a turbine bead mill.
3.2. Enzymatic Hydrolysis, Electrophoresis, and FTIR
After 6 hours of the enzymatic reaction, the degree of hydrolysis obtained on the protein extract was 84.84± 3.52%. While the biomass solution that was used as control (sample in phosphate buffer at the same reaction conditions without cell lysis) presented a degree of hydrolysis of 24.66 ± 1.05%.
The use of enzymatic complexes permits achieve more complete hydrolysis by overlaying the specific effects of each, as corroborate Morris et al. [17], who found degrees of hydrolysis greater than 50% in the enzymatic hydrolysis of Chlorella vulgaris biomass in suspension at 10 % (w/v) in water, with mixtures of papain-trypsin and papain-pancreatin.
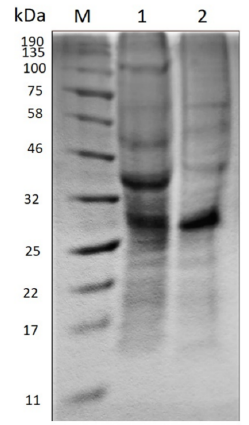
Polyacrylamide gel electrophoresis (SDS-PAGE) evidences the change in the molecular size of the proteins present in the extract before and after the enzymatic treatment as Figure 2 shows.
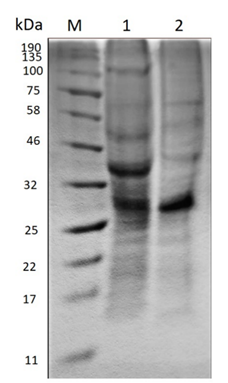
Source: The Authors.
Figure 2 Polyacrylamide gel electrophoresis (SDS-PAGE). M) Molecular marker. 1) Crude extract before hydrolysis. 2) Hydrolyzed extract.
In the crude extract before hydrolysis, the amount of bands existent suggests the presence of a large variety of proteins, the most prominent being between 25-32 kDa and 32-46 kDa. On the other hand, in the hydrolyzed extract the enzymatic action becomes evident when the molecular size of some proteins decreases, due to the diminution of the thickness of some bands such as that between 32-46 kDa and 46-58 kDa. Moreover, the band of approximately 100 kDa disappears and a new band stands out between 58-75 kDa.
Concerning the results of Fourier transform infrared spectroscopy (FTIR) analysis in Figure 3, the original biomass of Nannochloropsis sp. has an important peak between 2954-2850 cm-1 corresponding to the aliphatic chains of lipids and another one between 1710-1740 cm-1 related to esters of fatty acids and chlorophyll ketones [28].
Comparing the sample of the crude and hydrolyzed extract with the original biomass, a band of lower intensity between 3000 and 2850 cm-1 is presented and the band related to chlorophyll disappears, suggesting that the mechanical extraction process implemented removes very few amounts of compounds linked to lipids and pigments such as chlorophyll, being selective to proteins and carbohydrates.
The analysis by Fourier transform infrared spectroscopy, allows to relate the vibrational frequencies of the C = O and N-H bonds corresponding to Amida I and Amida II, with the secondary structures of the proteins, as a method to deduce the structural conformation of the proteins and the changes that could occur when some type of treatment is undergoing [29-31].
In the samples of crude and hydrolyzed extract, the typical peaks of protein secondary structures are evident [32,33]: β-sheets (1632 and 1637 cm-1), α-helices (1659, 1661 cm-1), β-turns (1683, 1681 cm-1), respectively.
Although the changes in the reported wavelengths and the intensities of the peaks attributed to the secondary structures were minimal, these could be because of the hydrolysis process is responsible for increasing the number of hydrogen bonds due to interactions between released species NH3 + and COO- with water, as Fang et al. deduced [29].
Apparently, this fact demonstrates that both the extraction protocol and the handling of the extracts during the different processes did not affect the folded structure of the proteins, maintaining their biological function [32,34].
3.3. Antimicrobial activity
Figure 4 shows the growth on the logarithmic basis of the microorganisms evaluated in the presence of three concentrations of extract in addition to an extract-free control sample.
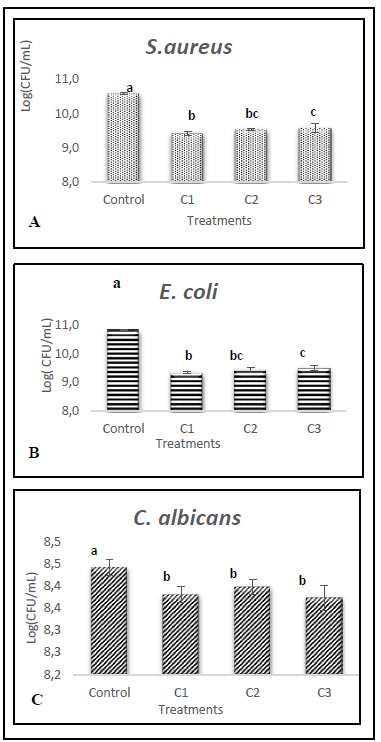
Source: The Authors.
Figure 4 Growth of A) S. aureus, B) E. coli and C) C. albicans with different concentrations of hydrolyzed extract (C1:33.35%(v/v); C2: 20%(v/v) y C3:10%(v/v)).
The results were analyzed using an analysis of variance (ANOVA) and Fisher's minimum significant difference test (LSD). The means of each treatment for the same microorganism with one letter in common do not differ significantly at 5%.
In the case of bacteria, there are statistically significant differences with a confidence level of 95% between the control sample and the samples containing the protein extract, as well as the extract samples with the highest concentration (C1: 33.35% (v/v)) and the samples with the lowest concentration (C3: 10% (v/v)).
In contrast, the growth of Candida albicans although there are statistically significant differences between the control sample and those where the protein extract was added, there are no differences between the samples with different concentrations of the extract.
Regarding the percentages of growth inhibition obtained for each treatment with different extract concentrations, the bacteria showed very similar inhibitions, nevertheless, E. coli was the microorganism most susceptible to its action, as shown in Figure 5.
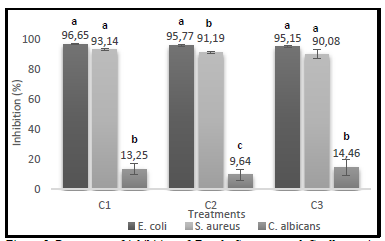
Source: The Authors.
Figure 5 Percentage of inhibition of E. coli, S. aureus and C. albicans in different concentrations of enzymatically hydrolyzed extract (C1:33.35%(v/v); C2: 20%(v/v) y C3:10%(v/v)).
The antimicrobial activity assay demonstrates that the hydrolyzed protein extract exhibits bacteriostatic activity against E. coli and S. aureus and fungistatic activity against Candida albicans.
The analysis of variance performed and Fisher's minimum difference test (LSD) shows the statistical differences between the microorganisms studied. The letters in common for each treatment do not differ significantly from 5%.
In all the treatments applied there were significant differences between bacteria (E. coli and S. aureus) and yeast (C. albicans). In the case of the extreme treatments, referring to the highest (C1) and lowest (C3) extract concentration, there were no statistically significant differences between the bacteria. On the contrary, in the treatment where the C2 extract concentration was applied, there were statistical differences among all microorganisms, being E. coli the most susceptible to its effect. This fact could be explained in part due to the production of proteases by S. aureus, which would block the action of the peptides present in the extract [35]. Sun et al. [11] presented similar results using a peptide fraction of the protein isolated from Spirulina platensis against E. coli and S. aureus, obtaining inhibition halos of 16 mm and 12 mm, respectively.
Other authors such as El-Baz et al. [12] evaluated a protein extract of Spirulina platensis that presented antimicrobial action against the microorganisms Enterococcus faecalis and Candida albicans but did not reveal any effect against the bacteria E. coli and Staphylococcus aureus.
4. Conclusions
The microalgae Nannochloropsis sp. is an important source of high-quality proteins. Due to enzymatic hydrolysis with a complex of papain-pancreatin proteases on the protein extract obtain this study, was achieved a degree of hydrolysis greater than 80%, reaching a considerable percentage of low molecular weight peptides (<3 kDa). These molecules showed significant percentages of inhibition on S. aureus, E. coli, and C. albicans, cataloging the extract as bacteriostatic and fungistatic, respectively, against the microorganisms evaluated.
Based on the results of the FTIR, apparently, the mechanical treatment implemented on the proteins of Nannochloropsis sp. did not affect the folding of the proteins and kept their biological function.