1. Introduction
The microbiota present in Colombia represents an unlimited source of biological potential as it includes microorganisms such as yeasts, fungi, and bacteria with great metabolic diversity. Particularly, filamentous fungi show great appeal in the area of biotechnology due to their easy cultivation [1], their potential to be used as edible strains, for the bioremediation of soils and water bodies [2] and their ability to generate a wide range of metabolites, including organic acids, antibiotics, and enzymes, which can be used in different types of industries [3]. That is why the importance and need to increase the use of these microorganisms in the medical, industrial or environmental sectors becomes evident, where direct benefits can be obtained from their use. One of the metabolites produced by fungi with the greatest industrial interest are enzymes. These have been important for the development of biotechnology-based processes, as they can eliminate the use of high temperatures, extreme pH values, organic solvents, and at the same time offer high substrate specificity, low toxicity, product purity and a reduced environmental impact [4]. Among the most used enzymes in the industry are: cellulases, which are complex and specialized enzymes that break down cellulose. This process has great applications in the paper industry as well as in the production of bioethanol from agro-industrial waste [5]; amylases, hydrolase enzymes that fulfill the function of catalyzing the hydrolysis reaction of the 1-4 bonds of the α-amylase component by digesting glycogen and starch, this makes them very useful in the food industry as well as for the processing of agroindustrial residues [6] and finally, ligninolytic enzymes that are capable of degrading lignin. It has been found that white-rot fungi produce these enzymes extracellularly, which makes them relevant in the paper industry and in the bioremediation field [7].
The type of metabolites that fungi produce is closely related to the place and substrate where they grow [8] and therefore, the analysis of their growth environment is key to understanding their role in the nutrient cycle and leads to a better understanding of the type of fungus collected. In addition, the frequency of appearance of a specific type of fungi gives information about the ecosystem where the collection was carried out. Hence, the importance of describing the conditions in which the samples are found, since all these variables in turn can help to determine the type of fungal communities that can be found in little-explored ecosystems [9] such as the Eastern Antioquia region [10,11] and provide a basis for recognizing other places where such communities may be located.
Thus, when the objective is the implementation of isolated fungi in industrial processes, their identification is of vital importance. Morphological characterization can be used to identify strains based on phenotypic characters. The limitation of this method is that it is not always possible to achieve a correct identification at the species level since morphological characters can often be misleading due to hybridization, cryptic speciation, and convergent evolution [12]. Hence, in recent years, molecular identification has been implemented as the main method to carry out the identification of fungi. In particular, the ITS region has been used to identify a large number of fungi with a high success rate [13]; even so, this region also presents problems for highly specious genera such as Aspergillus sp., Cladosporium sp., Fusarium sp., Penicillium sp., and Trichoderma sp. since their taxa have narrow or null gaps in the barcode of their ITS regions [12]. On the other hand, the LSU region is often used when phylogenetic analysis is desired to determine relationships between species since it turns out to be a molecular marker that can be useful for the placement of new fungal lineages from environmental studies in a comprehensive phylogenetic framework or for baseline fungal lineage analysis; it can even be useful for the identification of fungal species through the analysis of their D1 / D2 domain [14]. It has been reported that using this region, strains of filamentous fungi can be separated at the species level with a threshold of 99.81% [15], this region can be amplified and sequenced using the combination of the LROR and LR6 primers [12] to identify fungi found in ecosystems with great potential.
In this study, filamentous fungi were isolated from samples of wood and soil obtained from "La Lorena" natural reserve located in the municipality of Sonsón, Antioquia (Colombia). Then, using molecular identification and taxonomic keys, the strains found were characterized. This was made in order to determine the industrial application of the obtained fungi as an alternative for the sustainable use of the country's biodiversity, particularly by making use of the enzymatic potential of the isolated fungi.
2. Materials and methods
2.1. Collection and isolation of fungi
The strains were collected using a quadrant sweep in "La Lorena" natural reserve located in “Alto de la Sabana” district in Sonsón-ANT, Colombia (Fig. 1). Different substrates such as living or fallen trunks, litter and soil were examined in order to collect fruiting bodies and mycelium while soil samples were taken near the aforementioned substrates. Each sample was characterized according to macroscopic conditions such as color or appearance, substrate, place of growth, and location (latitude, longitude, height, temperature and humidity). The samples were preserved at low temperature in order to transport them to the laboratory of Universidad de Antioquia, Seccional Oriente. The collection of the samples was preceded by the permit for collection and mobilization of specimens (IDB 0321), processed before the National Environmental Licensing Authority (ANLA) by Universidad de Antioquia Research Vice-Principal’s office as a framework permit for the collection of species.
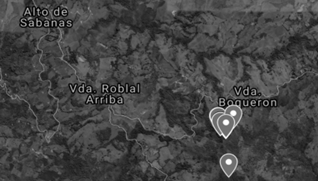
Source: the authors.
Figure 1 Satellite map of the sample collection area. The average collection conditions were: altitude 5°42, longitude -75°22, temperature 24.56 °C, altitude 2529 m and humidity 45.23 %.
Fruiting bodies and mycelium samples were cultured in a petri dish with PDA agar (Merck) using pieces of fungal tissue previously subjected to a disinfection process as described by Watling, 1981 [16].
In the case of soil samples, the protocol described by O'Donnell et al., 1992 [17] was followed, then 1 mL of each dilution was taken, and the sample was cultured as well in PDA agar (Merck). The agar used was not supplemented with antibiotics to inhibit bacterial growth. The cultured samples were incubated for a week at 30 °C and the growth of the strains was monitored.
The purification of the strains consisted of taking samples of the microorganisms that grew on the plate and culturing them using the streak plate method. The cultures were incubated again at 30 °C for one week and their growth was monitored. Those microorganisms in which it was possible to appreciate a single strain morphology growing on the PDA plate were considered as axenic cultures. Finally, the colonies with the macro and microscopic morphological characteristics of interest and of which there was a traceability over time were selected.
2.2. Macro and microscopic identification of fungi
The isolated strains were incubated for one week at 30 °C in 4 specific media: PDA Agar (Merck), Malta Agar (Scharlau), Sabouraud Agar (Scharlau) and Czapek Agar (Scharlau). The growth and macroscopic characteristics of the colonies such as coloration (front and back), shape, elevation, appearance, surface, margin or edge, and growth rate were monitored. The microscopic characteristics of the fungi were observed with the 40X and 100X objective using lactophenol blue [18] and the shape of the spores, hyphae (or mycelium) and reproductive structures were analyzed. To determine the genus of the specimen collected, the morphological characteristics were analyzed with taxonomic keys and identification literature of filamentous fungi [19-21]. This morphological characterization was subsequently corroborated with molecular techniques and sequencing.
2.3. Biochemical characterization associated with the enzymatic activity of fungi
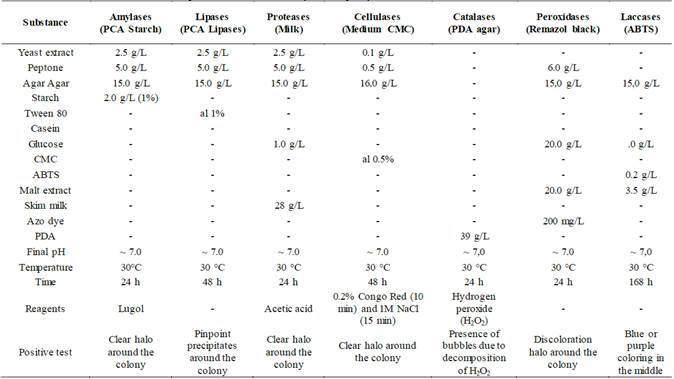
The biochemical characterization consisted of qualitative tests to evaluate the production of amylases, lipases [22], proteases [23], cellulases [24], catalases [25], peroxidases [27] and laccases [26]. The culture media used to induce production and determine the presence (+) or absence (-) of the enzymatic capacity of the isolated specimens, as well as the way of determining whether the test is positive or not, are specified in Table 1. Additionally, negative controls were made to have a reference point. To carry out these tests, the fungi were previously cultivated for a week at 30 °C on PDA agar (Merck) and from this culture a cut of approximately 5 mm of a piece of agar with mycelium of the fungus was made and transferred to each one of the media. The cultures were incubated at 30 °C for 3-4 days before developing the test.
2.4. Molecular identification of fungi
Selected fungi were molecularly identified by the amplification and sequencing of the LSU region of the 28S ribosomal DNA with the primers LROR, 5'-ACCCGCTGAACTTAAGC-3' (Tm = 54.29 °C) and LR6, 5'-CGCCAGTTCTGCTTACC-3' (Tm = 55.21 °C) [28]. The strains were cultured in triplicate in liquid PDA medium (400 g/L of potato, 20 g/L of glucose and 3 g/L of ammonium sulfate) and were incubated at 30 °C and 200 rpm for 3-4 days. The liquid culture was filtered to obtain the wet mycelium that was macerated to carry out the DNA extraction using the kit "Plant / Fungi DNA Isolation Kit # E5038" (NORGEN® Biotek Corp.) according to the manufacturer's protocol [29]. For the standardization of the PCR conditions, the conditions and quantities recommended for the enzyme "Taq DNA polymerase" (native, with BSA) # EP0072, stated in the kit, were used, which are shown in Table S1 and S2, available in the supplementary material.
Finally, the purification of the PCR products was carried out using the "GenElute Gel Extraction Kit" (Sigma-Aldrich) and using the manufacturer's protocol [30]. For each fungus, an electrophoresis of the PCR products obtained was carried out in a 0.8% agarose gel to facilitate the subsequent dilution of the gel; the bands obtained were cut with sterile blades. The concentration of the PCR products was measured in the Jenway Genova Nano spectrophotometer in order to ensure that the concentrations were in a range of 10 to 30 ug/mL, which is necessary to obtain good results using the Sanger sequencing offered by the Genewiz company. After eluted PCR products were sequenced, each sequence was subjected to a BLASTn search on the NCBI website [31]. Sequences that presented a query cover (“query cover”) ≥ 80% and a percentage identity (“percentage identity”) ≥ 97-100 [32] were considered as matches.
3. Results and discussion
3.1. Isolation and macro and microscopic identification of colonies
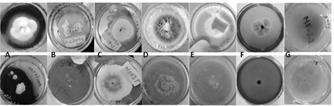
From the samples collected in the field, it was possible to isolate 13 filamentous fungi and 1 yeast in axenic culture using the described methodology. Among the strains identified based on their characteristics Trichoderma sp. genera (HF05.1, HF05.2, HF21, HF23, MF03.2), Penicillium sp. (HF09.2 and HF14), Mucor sp. (HF10 and MF05.1), Aspergillus sp. (SF05.1.1) and Lecythophora sp. (MF02.1) were found. In Fig. 2 the macroscopic and microscopic morphologies of the previously mentioned genera are shown.
The fungi that belong to the genus Trichoderma sp. (Fig. 2A-E), showed rapid growth and developed their characteristic morphology when cultured in PDA medium [33]. The colonies were characterized by having a velvety texture, presenting abundant sporulation and producing a yellow pigment in the agar.
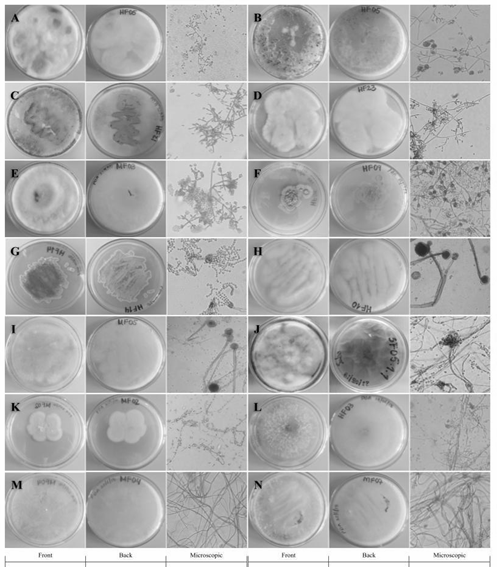
Source: the authors.
Figure 2 Macro and microscopic characteristics with 40X objective of: A) HF05.1, B) HF05.2, C) HF21, D) HF23 and E) MF03 .2 in PDA medium identified as Trichoderma sp.; F) HF09.2 in PDA medium and G) HF14 in Czapek medium identified as Penicillium sp.; H) HF10 and I) MF05.1 in PDA medium identified as Mucor sp. .; J) SF05.1.1 in Sabouraud medium identified as Aspergillus sp.; K) MF02.1 in PDA medium identified as Lecythophora sp.; L) HF03, M) MF04.2, N) MF07 in PDA medium without identification.
All the fungi had white mycelium and the spores had a greenish color except for HF23 (Fig. 2D). Under the microscope the characteristic structures belonging to the Trichoderma sp. genus such as septate and phialid solitary or grouped hyphae in the shape of a bottle with small and oval conidia were observed [20, 21].
In the case of Penicillium sp. (Fig. 2F-G), the colonies of both fungi presented a green coloration of different shades, with a rough texture. Under the microscope, globose conidia [19] and the brush-shaped conidiophore or “penicillus” characteristic of this genus [20] were observed, the appearance of which is favored due to the defined chemical composition of the Czapek medium [34]. The fungi belonging to the genus Mucor sp. (Fig. 2H-I) grew as white cottony colonies, with abundant aerial growth and turned brown (HF10) and black (MF05.1) with the passage of time. Coenocytic hyphae and common sporangia of this genus were observed under the microscope [20,21].
In the case of the isolate belonging to the Aspergillus sp. genus, (Fig. 2J), the growth of a white cottony colony that changed to green over time due to abundant sporulation was observed. Microscopically, the characteristic “aspergillaire head” was observed, consisting of the conidiophore, the vesicle, and the bottle-shaped phialides [20]. This structure could be easily observed thanks to the special composition of the Sabouraud medium [35].
Colonies of Lecythophora sp. (Fig. 2K) grew with a creamy salmon-pink texture and septate hyphae and oval conidia were observed under the microscope [36]. This yeast grows more regularly in media such as PDA and Malt, which stimulate the growth of this type of microorganism due to its balanced composition and high acidity [33,37].
The strains MF04, MF07 and HF03 (Fig. 2L-M), could not be identified with the culture media used since they did not present distinctive morphological structures that would allow a classification based on taxonomic keys that would relate them to already reported fungal genera. This may be due to the fact that the method for cultivating fruiting bodies has certain limitations related to the specific conditions they need to grow. Not to mention the bias of current culture media directed especially towards fast-growing species [38-40]. However, the substrates from which the fungi were collected, that is, decaying and living logs, litter and soil, support the type of fungi found [41-43].
3.2 Biochemical characterization associated with the enzymatic activity of fungi
The results obtained from the evaluation of the enzymatic capacities of the isolated strains are shown in
Table 2 and in Fig. S1, available in the supplementary material, both the positive and the negative results of the tests performed are observed.
Table 2 Results of the enzymatic capacities of the isolated strains.
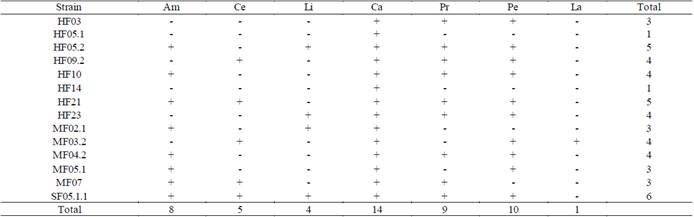
(+) shows activity, (-) shows no activity. (Am) amylases, (Ce) cellulases, (Li) lipases, (Ca) catalases, (Pr) proteases, (Pe) peroxidases and (La) laccases.
Source: the authors.
In Table 2, it can be observed how all the fungi showed catalase production, which is consistent considering that fungi are considered to be major producers of these enzymes since they are used to protect their cells from oxidative damage [25,44]. In contrast, only MF03.2 (Trichoderma sp.) fungus presented laccase activity, which indicates its potential for the production of ligninolytic enzymes associated with white-rot.
From the evaluation of the amylolytic capacity, eight fungi showed the presence of the activity (Table 2). The results obtained are consistent since amylases are usually associated with Aspergillus sp. genra (SF05.1.1) and Penicillium sp. (HF09.2), which coincide with the identified fungi; even so, other genera such as Mucor sp., Lecythophora sp. and Trichoderma sp., showed the presence of this activity.
Regarding the peroxidase capacity, in ten fungi the activity was evidenced (Table 2). Fungi HF05.2 (Trichoderma sp.), HF09.2 (Penicillium sp.), HF10 and MF05.1 (Mucor sp.) and SF05.1.1 (Aspergillus sp.) present a greater potential in the discoloration of remazol black. It should be noted that the mechanism by which discoloration occurs was not evaluated and the evaluation of this capacity was made taking advantage of the relatively nonspecific nature of ligninolytic enzymes that catalyze the oxidative breakdown of azo bonds [45].
Regarding the production of cellulases, five fungi belonging to Aspergillus sp. (SF05.1.1), Penicillium sp. (HF09.2) and Trichoderma sp. (HF21 and MF03.2) genera, presented the capacity to produce them. This enzymatic capacity gives indications of the type of fungus analyzed, since ascomycete fungi mostly produce enzymes responsible for the degradation of hemicellulose, soluble compounds and cellulose, substances present in the first stages of decomposition [42]. Even so, greater cellulase production was expected after having isolated multiple specimens characterized as Trichoderma sp.
The lipolytic capacity was the one with the lowest frequency of appearance, since it only appeared in four fungi (Table 2). The above finding is related to the type of substrates in which the fungi were collected, since the presence of fats in them is not predominant. The four samples that presented potential belong to Aspergillus sp. (SF05.1.1)., Trichoderma sp. (HF05.2, HF23) and Lecythophora sp. (MF02.1) genra.
Lastly, proteolytic capacity was present in nine fungi. The presence of this enzyme shows that the substrates where the strains were found also have proteins that serve as a carbon source apart from sugars and celluloses [46]. Among the genera found are Aspergillus sp. (SF05.1.1), Penicillium sp. (HF09.2) and Mucor sp. (HF10) and unlike what the literature reports, this activity was also predominant in isolates belonging to Trichoderma sp. (HF05.2, HF21 and HF23) genus.
HF05.2, HF21, and SF05.1.1 fungi tested positive for more of the enzymes evaluated in this study. These strains belong to the Trichoderma sp. and Aspergillus sp. genera [47]. It should be noted that the results of the presence of the activities mostly coincide with data previously reported in the literature.
Clearly, enzymes found are related to the substrates from which the strains were isolated and their presence indicates how fungi are capable of degrading different substances in order to use them as food sources. However, the amount and characteristics of the enzymes they produce vary according to specific environmental conditions, interactions, and the type of fungus. According to the above, more in-depth studies should be carried out with the fungi presented here to obtain a more complete characterization of the enzymes that are produced, also considering the particular characteristics of the strains, such as the relative ease of cultivation, which make them ideal for implementation in large-scale processes. Hence, the importance and impact they could have on industries such as: food (including clarification of juices and wines and brewing), biomaterials, cosmetics, detergents, energy (conversion of biomass into biofuels), agriculture, fine chemicals, food animal, leather, pulp and paper, pharmaceutical, textile, fat disintegration in wastewater treatment, mining and degradation of dyes or bioremediation [48].
In addition, along with the potential of using the strains to produce crude enzyme extracts, free enzymes or participate in biotransformation processes, it has been reported that these fungi produce other industrially important metabolites such as lactic acid, alkaloids, flavonoids, peptides, phenolic acids, phenols, quinones, steroids, terpenoids, among others, and are also capable of degrading polycyclic aromatics hydrocarbons [49]. Not to mention that the fungi of Trichoderma sp. genus are widely recognized for their potential to be used as biological control since they attack pathogens, which affect products important to humans.
Now, it is clear that the potential use of fungi in biotechnological applications begins with basic research on biodiversity and, therefore, the importance of this work, especially when currently not much development and sustainable alternatives are being generated using these microorganisms in regions with as much potential as the “Oriente Antioqueño,” this added to the growing interest in replacing synthetic products with more natural and eco-friendly products [50].
3.3. Molecular identification
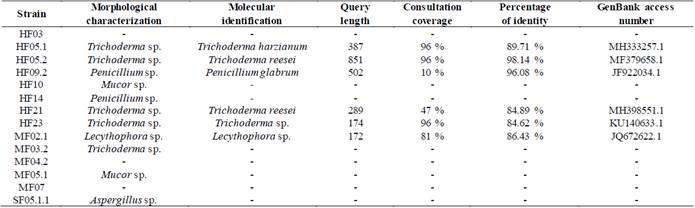
The agarose gels of the purified PCR products of the 28S gene (Fig S2, S3 found in the supplementary material) show that the size of the bands obtained with the DNA of the 14 isolates corresponds to the fragment size expected with the pair of LR0R / LR6(~1200 bp) primers [12]. Table 3 shows the results of the BLAST search and of the morphological characterization and molecular identification of the strains, showing that for HF05.1 (Trichoderma harzianum), HF05.2 (Trichoderma reesei), HF21 (Trichoderma reesei) and HF09.2 (Penicillium glabrum) strains, apart from verifying the genus of the fungi, it was possible to identify the species of these isolates and associate the access number to the Genbank, which shows how the use of molecular techniques is a key in this type of study [8] if it is considered that a correct species identification through morphological characterization is difficult due to the similarity of highly specious genera such as Trichoderma sp., Aspergillus sp., Mucor sp. or Penicillium sp. that have morphologically similar species [51] or the gap that exists within the nomenclature referring to the different sexual morphs of fungi [52]. Likewise, it is appreciated how by means of molecular identification the morphological characterization of the genus of HF23 (Trichoderma sp.) and MF02.1 (Lecythophora sp.) strains was corroborated.
Table 3 Results and coincidences between the identifications obtained in the morphological and molecular characterization.
Source: adapted from [2]
Additionally, Table 3 shows the values of the query length, which corresponds to the number of base pairs of the sequence to be studied, the coverage of the query, which is the value that represents how much of the sequence to be studied is covered by the target sequence and the percent identity that indicates how many characters in the two sequences are identical.
Taking into account the criteria to consider that a sequence made a “match,” only the results corresponding to the HF05.2 strain meet both conditions. For HF05.1, HF23 and MF02.1 strains, the consultation coverage values are in the required range but the percentage identity values do not reach the value of 97%, presenting instead values of 89.71, 84.62 and 86.43 % respectively. In relation to the HF09.2 and HF21 strains, both present relatively low coverage percentages with values of 10 and 47%, which indicates that, although the sequences have a percentage identity of 96.08% and 84.89%, the result obtained by BLAST may not fully match the species of fungus isolated.
It should be clarified that the region chosen to be sequenced (LSU) is usually used for phylogenetic analysis and to determine the relationships between species as it is a highly conserved region compared to the ITS region with which the identification of species is made because it is highly diagnosed [12]. This fact could explain the difficulty in achieving identification at the species level for the HF23 and MF02.1 strains. By virtue of this, the best thing for a correct molecular identification is to use both regions (LSU and ITS) or other molecular markers that allow making a contrast of the results [9].
This study also determined that for strains that were not identified with taxonomic keys or molecularly, data obtained with respect to biochemical characterization and morphological characteristics is key and allows a partial identification, even when these data are diverse and scattered and are not usually archived in databases that are integrated with sequence data [53].
4. Conclusions
The isolated strains that were collected in "La Lorena" natural reserve located in Sonsón-ANT, belong to Trichoderma sp., Penicillium sp., Mucor sp., Aspergillus sp. and Lecythophora sp. genera, with Lecythophora sp. being a pleomorphic yeast and the remaining filamentous fungi. These strains present enzymatic potential that make them suitable for being implemented in various industries, which is consistent with the ecosystem and substrates where they were extracted. Trichoderma sp. and Aspergillus sp. genera tested positive for more of the enzymes evaluated in this study. According to the above, the results obtained here offer a basis for the development of sustainable biotechnological processes focused on the degradation of industrial dyes and on obtaining extracellular hydrolytic enzymes of interest and commercial importance, using fungi present in the microbiota of Antioquia Eastern. Incidentally, if their potential is studied in depth, they could even be used in the fields of bioremediation and biological control.
Moreover, collecting and characterizing fungi from the environment, and even more if they are from non-studied habitats, represent a necessary tool to know their potential and in turn help to identify specimens and to evaluate the roles they have in the environment, which can contribute to the development of sustainable production processes. Consequently, it is imperative to continue with the study of the isolated specimens and increase efforts to feed the databases in order to identify a greater number of fungi since by not producing characteristic structures, their characterization is difficult. Therefore, studies of this type contribute to the recognition and evaluation of biodiversity and environmental potential of different regions, which facilitates future reseachs in the field of mycology that are directed to the growth and development of the country.