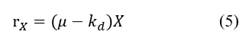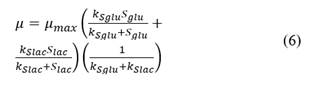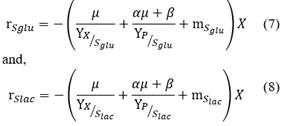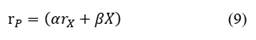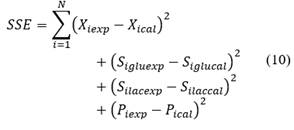1. Introduction
Lactiplantibacillus plantarum is an industrially relevant lactic acid bacteria (LAB) used in microbial food cultures (MFC) as a starter culture in the production of fermented food products such as yogurt, cheeses, sausages, pickles, and many others [1-3]. Furthermore, strains of L. plantarum can contribute to food safety by inhibiting pathogenic bacteria [4]. In other industries, L. plantarum is used to produce feruloyl esterase that has positive health benefits, such as promoting antioxidant, anti-inflammatory, and antimicrobial activity, since it is able to de-esterifying dietary fiber, and releasing hydroxycinnamates and derivatives during human and ruminal digestion [5]. Moreover, L. plantarum strains have been recognized as a Generally Recognized As Safe (GRAS) bacterium for humans [6,7].
Among the most important bottlenecks in L. plantarum production is its growth in a bioreactor. In this case, the bacterium is exposed to several influencing factors including culture medium, pH, temperature, agitation, final products formation, etc. [8]. The culture medium for biomass production must be optimized in order to improve the yields (YX/S, YP/S, YP/X) and reduction of production cost [3]. We proposed a co-substrate (glucose and lactose in whey as carbon source), since dairy industries generate a considerable amount of whey as by-product which, without an appropriated treatment could be a source of environmental contamination due to its high biochemical oxygen demand [9]. Moreover, the co-substrates can be satisfactorily used in biotechnology process. For example, the production of xylitol from xylose and glucose, as the co-substrate, using Candida moggi ATCC 18364 yeast, allowed an increase in the specific growth rate, biomass yield, and volumetric productivity of xylitol compared with the production of xylitol using xylose as the sole carbon source [10]. In another study, the production of xylitol by Debaryomyces nepalensis NCYC 3413 strain, was influenced by using a co-substrate (glucose and/or arabinose). They achieved high xylitol yield by using xylose and glucose as co-substrate in a ratio of 9:1 respectively [11].
L. plantarum produces lactic acid as the major metabolic end-product of carbohydrates [12], and the lactic acid produced inhibits the rate of cell growth. In order to improve the production of cell growth (biomass), methods for measuring, monitoring and modelling bioprocesses have been developed [13]. In particular, mathematical models can be used to control and optimize bioprocesses [14,15]. These mathematical models are used to explain the relationship between kinetics of cell growth, substrate consumption, and product formation, so, helping to predict parameters such as the conversion rate and yield [7]. Several classification criteria for mathematical models have been used. One of them classifies mathematical models as structured or unstructured. Structured models describe individual cellular processes and reactions by linking to specific RNA, DNA, proteins, etc., with the objective to obtain a more realistic description; whereas, unstructured models are mainly used in order to describe bacterial kinetics in complex natural substrates [16,17].
Some mathematical models for cell growth of LAB have been reported [18-20]. Regardless of the great variety of growth and production models proposed for LAB, these models are directly applicable to cell growth and lactic acid production when two substrates are simultaneously consumed. With the goal to establish an accurate mathematical model for co-substrate fermentation of L. plantarum 60-1, a modification of the basic Monod equation by including a product inhibition term, is proposed in the present work [7]. To verify the model, batch fermentation experiments were conducted using a LAB strain recently isolated in our laboratory, L. plantarum 60-1, which consumes both glucose and lactose [21].
2. Materials and Methods
2.1. Bacterial Strain
The bacterial strain L. plantarum 60-1 used in this study was isolated and identified from the Colombian sour cream called “Suero costeño” in our previous work [21]. The strain was stored in the culture collection of Biotransformación research group (University of Antioquia, Medellín, Antioquia, Colombia). The strain was cryopreserved in 2 mL vials at -20 ºC in Man Rogosa and Sharpe broth (MRS; Oxoid, Ltd., Basingstoke, UK) with 30% (v/v) glycerol (PanReac Química SLU, Barcelona, Spain) [21].
2.2. Batch culture conditions
Batch culture experiments were carried out in a 5 L, stirred tank reactor, with a 4 L working volume (BioFlo 115, New Brunswick, USA). The temperature was automatically controlled at 32( 1 ºC, impeller speed was 100 rpm, pH was kept at 5.5( 0.5 by addition of 5.0 NaOH (Merk, Darmstadt, Germany) [22], and foam formation was controlled automatically by the addition of a silicone antifoam sterile solution (4% v/v, Sigma Aldrich, USA).
The Bio-1 culture medium, used in batch cultures, contained glucose (20 g/L), lactose in whey (10 g/L), yeast extract (4.7 g/L) ammonium sulfate [(NH4)2SO4](3.3 g/L), magnesium sulfate [MgSO4] (0.2 g/L), manganese sulfate [MnSO4] (0.05 g/L), monopotassium phosphate [KH2PO4] (0.2 g/L) and sodium citrate [Na3C6H5O7] (2 g/L). The pH of the culture medium was 5.5( 0.5. The composition of whey was 72.6% lactose, 12.4% protein, 1.0% fat, and 5.16% ash. All components used in the preparation of culture medium were reagent grade, except for glucose, whey, and yeast extract which were USP grade.
The inoculum volume was 10% (v/v) of the total work volume and it was prepared as follows: two milliliters of the cryopreserved strain (1 x108 CFU/mL, stored at -20 ºC) were thawed at room temperature and used to inoculate 40 mL of MRS broth in a 100 mL flask, followed by incubation with shaking at 100 rpm and 32 ºC for 16 h (1st generation). The culture was then aseptically transferred to 500 mL flask with 360 mL of Bio-1 broth and incubated at 100 rpm and 32( 1 ºC for 5 h (2nd generation), which was used as the inoculum. The batch culture experiments were conducted at 100 rpm and 32( 1 ºC ºC in the 5 L reactor for up to 24 h. Samples were withdrawn at intervals of 1 h for the first 10 h and a final sample was withdrawn at 24 h.
2.3. Analytical methods
Biomass concentration was determined by the dry weight method [23]. A volume of 1.5 mL sample was centrifuged for 15 min at 4.722 g. The pellet formed was dried in an oven at 80 ºC for 48 hours (until constant weight). The biomass (g/L) was calculated with the difference in weight between the sterilized Bio-1 broth without inoculation vial and the vial with the dry sample. To measure vial weight, an analytical balance was used (Sartorius AG, Goettingen, Germany).
Glucose, lactose, and acid lactic concentrations were analyzed in a high-performance liquid chromatography (HPLC) system. The chromatograph used was Agilent Technologies 1200 (California, USA), model 61362A with an ionic exchange separation column HPX-87H, 300×7.8 mm. The samples were centrifuged for 5 min at 4.722 g, and the supernatant was diluted in the mobile phase H2SO4 (0.008 N). Every diluted sample was filtered using 0.2 μm regenerated cellulose filters. After that, the sample was injected under the following running conditions: 20 μL sample volume, 0.6 mL/min flow, 35 °C column temperature and 12 minutes runtime.
2.4. Kinetic models
There are several unstructured mathematical models used in biotechnology which describe growth kinetics of LAB [24-26]. We have used a model which includes two carbon sources (glucose and lactose) for the growth of L. plantarum 60-1 and lactic acid production.
Differential mass balance equations describe biomass growth eq. (1), co-substrate consumptions eq. (2 and 3), and lactic acid production eq. (4) in a batch model of L plantarum 60-1.
The mass balance for biomass is described as follows:
where r X is the volumetric rate of biomass formation.
The mass balances for glucose (S glu ) and lactose (S lac ) substrates are described as follows:
where, r Sglu and r Slac are the substrate consumption rate.
The mass balance for lactic acid production (P) can be described by the Luedeking-Piret equation [27] as follows:
where, r P is the volumetric rate of lactic acid production.
The kinetics with double Monod relation, constant yield relation (for glucose and lactose) and product formation are shown in eqs. (5)-(9).
where μ is the specific growth rate, and k d is the specific death rate. As the specific growth rate (μ) is higher than the specific death rate (k d ) then the value for this parameter is assumed zero (μ >>k d ), besides, this model is non- segregated. We have used double Monod kinetics, for two carbon sources (glucose and lactose) as substrates. The constitutive equation (μ) considering parallel reactions is described as follows:
where, k
Sglu
and k
Slac
are the Monod constants for glucose and lactose respectively [16]. Eq. (6) gives an additive, fractional contribution for each substrate. Each substrate allows a different maximal growth rate. To describe the effects of substrate limitation on the specific growth rate, the term

were included.
where, r Sglu and r Slac are the substrate consumption rate, Y X/S is the cell yield coefficient, Y P/S is the product yield coefficient, m Sglu and m Slac is the cell maintenance coefficient for glucose and lactose respectively. In order to avoid negative substrate concentrations, a conditional in the model was employed. For substrate concentrations less than zero the substrate concentration becomes zero.
where, r P is the volumetric rate of lactic acid production, ( is the growth-associated product constant and ( the non-growth associated product constant.
The simulation was carried out in Berkeley Madonna© software (version 10.2.8 for macOS). Four ordinary differential equations were solved simultaneously using the fourth order Runge-Kutta method. The data were statistically analyzed calculating standard deviation and Global relative error (Microsoft® Excel for Mac Version 16.35 (20030802) [28].
2.5. Experimental design
A total of six experiments were carried out in order to determine the model parameters identification- MPI (n=2) and the model validation- MV (n=4). The conditions employed for biomass, glucose, and lactose initial are shown in Table 1. Descriptive analysis of the biomass, glucose, lactose, and lactic acid concentrations was obtained from two independent trials and each analysis was made in triplicate. The diagram of the experimental procedure in the present study is shown in Fig. 1.
Table 1 Initial conditions for the experiments in a 5 L stirred tank reactor. Values for model parameters identification (MPI) and the model validation (MV).
| Target | Biomass initial (g/L) | Glucose initial (g/L) | Lactose initial (g/L) |
|---|---|---|---|
| MPI1 | 0.3 | 20 | 10 |
| MPI2 | 0.3 | 20 | 10 |
| MV1 | 0.3 | 10 | 10 |
| MV2 | 0.3 | 10 | 5 |
| MV3 | 0.3 | 4 | 8 |
| MV4 | 0.3 | 5 | 5 |
Source: The Authors.
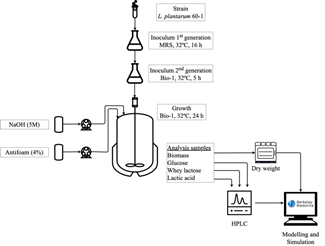
Source: The Authors
Figure 1 Diagram of procedure for experiments and modeling of growth, substrate consumption and lactic acid production by L. plantarum 60-1.
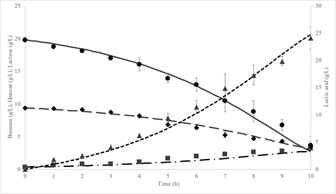
Source: The Authors
Figure 2 Experimental data and kinetic model simulation of L. plantarum 60-1 in a batch culture: Biomass model (dash-dotted line), biomass experimental (squares), glucose model (solid line), glucose experimental (circles), lactose model (dashed line), lactose experimental (diamonds), lactic acid model (dotted line), lactic acid experimental (triangles).
2.6. Parameter estimation
Due to the relationship between different data sets of the kinetic from L. plantarum 60-1, the accuracy of the parameters in the model can be increased by incorporating this relationship in the fitting process, instead of fitting the recordings separately [29]. An algorithm to fit multiple measured curves simultaneously was used in order to identify the parameters. The software used was Berkeley Madonna© (version 10.2.8 for macOS). The range of values (minimum to maximum) were taken from the literature. Berkeley Madonna© software minimizes the sum of the square deviation between data output of the model and experimental data (dataset). The parameter estimation is the sum of squared errors for each variable state (biomass, glucose, lactose and lactic acid). The sum of all terms is:
2.7. Model validation
The model was validated (MV) by using the average of four fermentation batchs that were not used for estimating the model parameters. For the model validation, different experimental sets of initial concentrations of biomass, glucose and lactose were performed. Finally, the global relative error was calculated.
3. Results and Discussion
3.1. Growth, co-substrate consumption and lactic acid production by L. plantarum 60-1 strain
Average and standard deviation of experimental data, as well as, kinetic model simulating the bacterial growth, co-substrate consumption and lactic acid production, are shown in Fig 2. The exponential phase started immediately after inoculating the bioreactor with an inoculum concentration of 0.34 ± 0.1 g/L.
During the growth of L. plantarum 60-1, lag phase was not observed. The lag phase indicates the adaption of cells to their new environment [30]. The lack of lag phase in the bioreactor could be due to the process of obtaining the inoculum. The inoculum was obtained in two steps in the present study. The first step called “first generation” occurs from inoculation of the cryopreserved strain in MRS broth and incubation for 16 h. The second step called “second generation” occurs since the culture in MRS broth was transferred to the Bio-1 broth, followed by incubation for 5 h. The transition between MRS into Bio-1 broth, which is the same culture medium used in the bioreactor, before inoculation in the bioreactor may have avoided the occurrence of the lag phase. Similar to our results, Atehortúa et al. [15] did not observe the lag phase in the cellular growth when the authors obtained the inoculum for bacterial growth in two steps, changing the culture medium from laboratory reagents grade to USP grade; the latter was used in the pilot scale fermentation. Our results differ from Alvarez et al. [31] who characterized the kinetics of biomass and lactic acid productions, as well as substrate consumption of Lactobacillus casei var. rhamnosus from Sacco-Clerici (currently renamed as Lacticaseibacillus rhamnosus) [32] cultured in deproteinized whey. The authors found lag phases in the range of 5 to 10 h because they used freeze-dried inoculum in one step.
The growth of L. plantarum 60-1 in Bio-1 culture medium had a significantly increased the biomass from 0.34 (0.1 to 3.42 (0.08 g/L after 24-h fermentation. Sharma et al. [33] compared the L. plantarum NCDC 414 growth in MRS broth and vegetable juice and found a growth from 0.0315 g/L to over 2.1 g/L in the MRS broth, but a smaller growth increase from 0.01 g/L to over 1.328 g/L for vegetable juice. Thereby, non-conventional culture medium also allows the cell growth in a pilot scale bioreactor in order to satisfy nutritional requirements of the culture at a low cost.
The concentration of glucose and lactose was initially 19.83 (0.41 g/L and 9.39 (0.25 g/L, respectively. At the end of the fermentation, there was a consumption of 99.75% for glucose and 91.16% for lactose. Both glucose and lactose limited the overall cell-growth rate. However, the final concentration for glucose was 0 g/L, while that of lactose was 0.83 and 0.87 g/L for model and experimental data respectively.
Former genus Lactobacillus has a broad ecological distributions and diverse habitats, since, given its metabolic flexibility, is able to utilize a wide range of carbohydrates; however, it does not grow on the pentoses, as it lacks of fructose-1,6-diphosphatase which plays a key role in gluconeogenesis [34]. A concomitant consumption of glucose and lactose by L. plantarum 60-1 was observed in the present study, which differs from common knowledge that lactobacilli rarely uses different carbon sources simultaneously [35]. To achieve maximal growth, lactobacilli organize the carbohydrate consumption in a hierarchical way in which the utilization of carbon source begins with the preferred one, until it is exhausted, and then the second carbon source begins to be consumed, i.e., a diauxic growth [36].
Via the Embden-Meyerhof pathway, LAB including L. plantarum strains can metabolize hexoses to produce lactic acid [19, 37]. LAB contain two systems for the transport of lactose (disaccharide) into the cell facilitated by phosphoenolpyruvate (PEP): lactose phosphotransferase system and lactose permease system [38, 39]. The concomitant consumption of glucose and lactose found in L. plantarum 60-1 can be explained by the carbon catabolite repression (CCR) mechanism. CCR is used in bacteria as a mechanism which controls if different carbon sources are metabolized in parallel or sequentially [40, 41]. This is the first report of L. plantarum 60-1 where a co-substrate system different from the diauxic growth was used. Similar to our study, Bartkiene et al. [4] used an alternative substrate for the growth of L. plantarum LUH135 in a medium composed of whey enriched with glucose (2.5%), yeast extract (2.0%) and sucrose (0.5%); however, they did not present the kinetics of substrate consumption unlike our results.
Lactic acid is a metabolic product directly associated with energy generation pathways in LAB [42]. Fig. 2, shows the lactic acid production kinetics. Lactic acid was produced at the beginning of the fermentation, after 10 h the value was 24.09 (1.95 g/L and its final value (24 h) was 31.67 (1.14 g/L and 31.60 g/L for experimental data and simulation, respectively.
As the lactic acid production by L. plantarum 60-1 begins from the early stage after the inoculation of bioreactor, the pH tends to drop, as reported in previous work; the pH dropped to 3.6 with residual glucose concentration of 1.32 g/L [43], and the cell growth can be inhibited. However, cell growth inhibition was not observed in the present study, possibly because pH was maintained at 5.5±0.5. Similarly, to our results, Fu et al. [1] found that batch fermentations of L. plantarum ATCC 21028 without pH control resulted in pH lower than 4.0 and inhibition of the cell growth; when pH was controlled, the optimum pH for the strain was found to be between 5 and 6. In the same way, Mechmeche et al. [18] evaluated the growth of L. plantarum isolated from traditional fermented olive in MRS broth. They found a maximum biomass concentration and lactic acid production of 2.02 g/L and 4.32 g/L, respectively, in a process without pH control, with the pH decreasing to 4.75 and the final concentration of sugar reaching 6.82 g/L. The authors suggested that the growth was limited by lactic acid production instead of substrate limitation, and reported an inflection point (stationary phase) after 8 hours of fermentation [18]. In a similar study, Passos et al. [19] developed an unstructured batch model to describe bacterial growth, lactic acid production, and substrate consumption by L. plantarum MOP3 in cucumber juice without pH control. The initial and final sugar concentrations were 12.96 g/L and 5.06 g/L, respectively, the final biomass concentration was 0.5 g/L, and the pH dropped to 3.2 [19]. Another study by Luedeking et al. [27] found the drastic influence of pH on yields and rates of growth, substrate consumption and product formation of L. acidophilus, with the growth being faster when the pH was controlled than when it was not controlled.
For this reason, controlling pH is important in fermentation processes when acid is produced. Gupta et al. [44] carried out the growth of L. plantarum strain on raw and heat-treated forms of seaweeds without pH control, the pH was reduced from 6.6 to 3.9 due to the amount of organic acids produced (2.5 g/L). In the same way, Zacharof et al. [7] modelled and simulated L. lactis strain in three different production systems and found that, without pH control, complete growth inhibition did not occur, but the produced lactic acid decrease biomass formation. These studies indicate that L. plantarum grows better in slightly acidic culture media with an initial pH between 5.4 and 6.4 but the growth is commonly stopped when a pH 3.6- 4.0 is reached, depending on the species and strain [34].
3.2. Batch kinetics model of L. plantarum 60-1
The set of model parameters for batch culture of L. plantarum 60-1 is shown in Table 2. The parameters associated to biomass growth were μ and k d . The specific growth rate has the following parameters associated μ max , k Sglu , k Slac , Y X/Sglu , Y X/Slac , Y P/Sglu , Y P/Slac , m Sglu , and m Slac . The value of μ max was 0.061 h-1, which is less than other L. plantarum strains with reported value being between 0.2 and 0.4 h-1 [1,18]. As we used glucose and lactose as two different carbon sources, the overall reaction growth rate was lower than the rate expected, due to double substrate limitations instead of a single substrate limitation [45].
Table 2 Set of model parameters obtained from batch cultures of L. plantarum 60-1
| Name | Symbol | Value |
|---|---|---|
| Maximum growth rate (1/h) |
|
0.061 |
| Monod constant for glucose (g/L) |
|
4.400 |
| Monod constant for lactose (g/L) |
|
2.260 |
| Yield of biomass from substrate-glucose (g/g) |
|
0.280 |
| Yield of biomass from substrate- lactose (g/g) |
|
2.130 |
| Yield of product from substrate- glucose (g/g) | > |
3.050 |
| Yield of product from substrate- lactose (g/g) |
|
5.000 |
| Maintenance coefficient for glucose (g/g·h) |
|
0.021 |
| Maintenance coefficient for lactose (g/g·h) |
|
0.014 |
| Growth- associated product constant (g/g) |
|
10.070 |
| Non-growth associated product constant (g/g·h) |
|
0.0058 |
Source: The Authors.
Two mass balances for glucose and lactose consumptions were applied in a batch process (Eq. 3 and 4). The equations for substrate consumption include the use of substrates in different metabolic pathways. Substrate consumption is used for growth (r X /Y X/S ) and product synthesis (r P /Y P/S ), while another fraction is used to generate energy for maintenance activities (m s ). These different parameters are shown in Table 2 and can be related using the yield and maintenance parameters. However, the value of m s was determined to be less than 0.05 g/g h in the present study, which is similar to the value (0.057 g/g h) reported by Zacharof et al. [7].
The lactic acid production rate (r P ) is related to the L. plantarum 60-1 growth rate (r X ) and the bacterial density (X), as expressed Equation (4), as lactic acid production must account for growth associated production (() and non-growth associated production ((), with the estimated parameters shown in Table 2. According to the results, the growth rate of biomass production was not inhibited by acid lactic production but, instead, was inhibited by substrate consumption [30].
Between the 10-24 hours lactic acid concentration increased by 21.79%, which means that although the growth is generally the major energy-demanding functions of cells, the lactic acid production is coupled to energy metabolism during cell growth [46]. Furthermore, the parameters ( and (, for the associated and non-growth associated product, respectively, indicate that lactic acid is produced in exponential and stationary phases of the batch growth, with most produced in the exponential phase (Fig. 1) [30]. Similar to our results, Altiok et al. [47] developed a kinetic model for lactic acid production from whey by L. casei NRRL B-441; however, they used the logistic equation with biomass and product inhibition terms by modifying the equation of specific growth rate.
3.3. Model validation of L. plantarum 60-1
The information about the average of initial and final concentration of biomass, glucose and lactose for the experiments (MPI and MV) are shown in Table 3.
Table 3 Concentrations initial and final of the variables (X, S Glu , S Lac and P) for the experiments in a 5 L stirred tank reactor. Values for model parameters identification (MPI) and the model validation (MV).
| Target | MPI | MV1 | MV2 |
|---|---|---|---|
| Biomass initial (g/L) | 0.39± 0.1 | 1.13± 0.24 | 0.38± 0.21 |
| Glucose initial (g/L) | 19.83± 0.41 | 10.97± 0.07 | 4.40± 0.16 |
| Lactose initial (g/L) | 9.39± 0.25 | 10.51± 0.11 | 7.45± 0.23 |
| Lactic acid initial (g/L) | 0 | 0 | 0 |
| Biomass final (g/L) | 2.84± 0.05 | 2.22± 0.35 | 2.12± 0.15 |
| Glucose final (g/L) | 0.05± 0.07 | 0 | 0 |
| Lactose final (g/L) | 0.83± 1.17 | 0 | 0 |
| Lactic acid final (g/L) | 31.67± 1.14 | 20.7± 2.31 | 11.91± 0.14 |
Source: The Authors.
The validation experiments demonstrate the robustness of the kinetic model. The results of model validation are shown in Table 4.
4. Conclusions
This is the first report where modeling kinetics of lactic acid production, co-substrate consumption, and biomass obtaining by L. plantarum 60-1 strain was investigated. The cell growth did not show lag phase, and the lactic acid production started from the beginning of the growth, which is desirable in order to reduce the duration of the fermentation. The biomass growth rate declined after 10 hours of the fermentation due to the consumption of the growth-limiting substrate, leading to stationary phase. The consumption of glucose and lactose was concomitant, and the accurate control of pH at 5.5 enabled the consumption of 99.75% glucose and 91.16% for lactose at the end of fermentation. The experimental values enabled the fitting of the modified kinetic model. The validity of our work in modifying the Monod equation by including co-substrate consumption, without including the product (lactic acid) inhibition term. The model developed for a batch process could be adapted for other operation modes with the same microorganism, in order to improve the understanding of the fermentation process with L. plantarum 60-1 strain.