Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Colombiana de Ciencias Químico - Farmacéuticas
Print version ISSN 0034-7418
Rev. colomb. cienc. quim. farm. vol.41 no.1 Bogotá Jan./June 2012
Artículo de investigación tecnológica
Development and validation of a TaqMan multiplex PCR assay for the Gene Dosage Quantification in cancer
Desarrollo y validación de PCR múltiplex con sondas TaqMan para la cuantificación de la dosis génica en cáncer
Sandra Janeth Perdomo Lara1,2, Sandra Johanna Morantes1, Fabio A. Aristizábal Gutierrez1
1 Grupo Farmacogenética del Cáncer, Departamento de Farmacia, Facultad de Ciencias, Universidad Nacional de Colombia, carrera 30 Nº 45-03, Bogotá, D. C., Colombia. Corresponding author, e-mail: faaristizabalg@unal.edu.co.
2 Facultad de Odontología, Unidad de Investigación Básica Oral (UIBO), Universidad el Bosque, carrera 7B Bis Nº 132-11, Bogotá, D. C., Colombia.
Recibido para evaluación: 10 de abril de 2012.
Aceptado para publicación: 19 de junio de 2012.
SUMMARY
Gene dosage tests are very important for the molecular diagnosis of diseases caused by either deletion or amplification of a specific DNA region containing certain genes. Changes in gene copy number may lead to under- or over expression of genes responsible for the disease phenotype. Discovering these defects and understanding their biological meaning can lead to improved therapeutic opportunities in cancer. Fluorescent in situ hybridization (FISH) still remains the gold standard method for gene dosage analysis. However have several limitations since locus specific probes are expensive, the procedure is significantly time-consuming and tandem microduplications may be undetected as a result of the limited resolution on metaphase spreads. Quantitative Real-time PCR is a rapid assay with results available in 24h, has advantages in terms of sensitivity and specificity. In the present study, we have developed an assay using TaqManTM multiplex Real-time PCR for gene dosage analysis of oncogenes EGFR, ERBB2, AKT2, CMYC, MYCN, MYCL1, PI3KCA and REL in human cell lines. This method calculates the copy number of each oncogen and is a promising alternative technique to FISH and Southern blot. Therefore, this technique could be considered as a powerful method gene dosage quantitation in clinical and research genetic surveys.
Key words:: cancer, oncogenes, gene dosage, real time PCR, TaqMan probes.
RESUMEN
La evaluación de la dosis génica constituye una herramienta importante para el diagnóstico molecular de enfermedades causadas tanto por la pérdida o amplificación de una región específica de ADN. El cambio en el número de copias génicas, puede conllevar a la pérdida o sobreexpresión de los genes responsables de fenotipo de la enfermedad. El descubrimiento de estas alteraciones y la comprensión de su significado biológico pueden conducir al incremento de las oportunidades terapéuticas en cáncer. La hibridación fluorescente in situ (FISH) es el método de referencia para el análisis de la dosis génica amplificación. Sin embargo, presenta algunas limitaciones relacionadas con el costo de las sondas locus específicas; el procedimiento es demorado y las microduplicaciones en tándem podrían no ser detectadas como resultado de la limitada resolución de las metafases. En este sentido, la PCR cuantitativa es una metodología rápida y tiene ventajas en términos de sensibilidad y especificidad. En el presente estudio, se estandarizó la PCR multiplex en tiempo real para el análisis de la dosis génica de los oncogenes EGFR, ErbB2, AKT2, cMYC, MYCN, MYCL1, PI3KCA y REL en líneas celulares tumorales humanas, como una técnica alternativa al FISH para evaluar la dosis génica en muestras clínicas de cáncer.
Palabras clave cáncer, oncogenes, dosis génica, PCR en tiempo real, sondas TaqMan.
INTRODUCTION
In mammals, genetic alteration in gene copy number by amplification or duplication has been defined as one of the common mechanisms that lead to deregulation of gene expression and to neoplasic transformation (1-3). The number of copies of a gene per haploid genome (gene dose) has been analyzed traditionally by techniques such as Southern blot or fluorescent in situ hybridization (FISH). However, these techniques may not be appropriate when working with small amounts of starting material (i. e., paraffin embedded tissues, free plasma DNA) (9, 10).
The accurate determination of the number of copies of a gene in the genome (gene dosage) is essential for a number of genetic analyses. Quantitative real time PCR (qPCR) detection has shown advantages over traditional Southern-blot and FISH techniques.
Recently, a real-time PCR assay using a TaqMan probe (a fluorescent DNA probe based on the 5´ to 3´ exonuclease activity of the Taq polymerase) has been developed for quantitative DNA analysis (6). The oligonucleotide probe, with a reporter fluorescent dye attached to its 5' end and a quencher dye attached to its 3´ end, hybridizes to the target gene. During PCR amplification, the quencher dye is cleaved by the 5´ nuclease activity of Taq polymerase resulting in the accumulation of reporter fluorescence. The release of the fluorescent dye during amplification allows for rapid detection and quantification of DNA (14). Compared with other methods the TaqMan real-time PCR assay has advantages in terms of sensitivity, rapidity and specificity.
Genetic gains and losses, i. e. changes in gene dosages, are common abnormalities of human cancers that regulate the expression of genes and are motive forces for this evolution (1). Tumor cells bearing an increasing number of gains and losses successively emerge and are selected for based on the growth advantage caused by the genetic changes. Discovery and functional assessment of gene dosage alterations involved in carcinogenesis are therefore essential for understanding the biology of the disease and can lead to improved therapeutic opportunities. The purpose of the present work was to design qPCR multiplex assay with TaqMan probes detection to determine the gene dosage of oncogenes EGFR, ERBB2, AKT2, CMYC, MYCN, MYCL1, PI3KCA and REL in tumors human cell lines as molecular markers for cancer.
MATERIAL AND METHODS
Samples and DNA extraction
The gene dosage analysis was performed in cell lines obtained from ATCC (American Type Culture Collection): HEp-2 (human laryngeal carcinoma), PC3 (prostatic adenocarcinoma), HepG2 (hepatocellular carcinoma), Hep3B (human hepatoma), MCF7 (breast adenocarcinoma), MDA-MB (breast adenocarcinoma), HT29 (colorectal adenocarcinoma) and MRC5(normal Lung Fibroblasts), A549 (Human lung adenocarcinoma epithelial cell line), H727 (bronchial carcinoid), H292 (lung, carcinoma, mucoepidermoid), H520 (lung squamous cancer cell line), H82 (small cell lung cancer) and the cell line SPG2 from a Colombian patient with large cell anaplastic carcinoma. The cells were grown in culture medium supplemented to reach confluence. The DNA of the cell lines were obtained by enzymatic digestion with proteinase K (2 mg/ml), extracted with phenol-chloroform isoamyl alcohol and ethanol precipitation. We assessed the quality of the extracted DNA by conventional PCR to amplify a 171 bp segment of the B-actin (ACTB) gene and agarose gel electrophoresis.
TaqMan multiplex PCR assay for Gene Dosage Quantification
The number of gene copies in each sample was evaluated by real-time multiplex PCR using Taqman probes. For this, two PCR mixes were performed to amplify 1. EGFR, ERBB2, AKT2; 2. MYC, MYCN, MYCL1 y 3. PI3KCA, REL and ACTB genes. The gene dosage of each oncogene was determined, utilizing the gene ACTB as reference, with the double-delta CT relative quantification (2-Δ ΔCT) method (6) in duplicate assays, using the kit TaqMan® Fast Universal PCR Master Mix in the Applied Biosystem 7500 instrument. The calculations were made according to manufacturer's specifications.
It was confirmed earlier that the efficiencies of the amplifications for each gene were similar, using the method published by Kenneth et al. (19).
The reactions were made in a final volume of 25 µl, containing: 1x Master mix, 0.04 µM of each primer and 0.375 µM of each probe. The PCR program used was: 95oC x 10 min, 50 cycles (95oC x 15 sec, 61oC x 1 min). Table 1 shows the sequence of primers and probes used. A 2-Δ ΔCT value greater than 2 was considered gene amplification. The change in target gene copy number was calculated for each gene and sample using the double delta formula (2-Δ ΔCT), where Δ ΔCT = (Cttarget - Ct-ACTB) CANCER - (Cttarget – Ct-ACTB) NORMAL. A > 2 2-Δ ΔCT value was considered a significant gene dose increase (6). The change in gene dosage in the cancer cell lines was compared with the gene dosage of normal lung fibroblasts cell line (MRC5).
To the left of each probe is mentioned the fluorochrome that was used as the "quencher" and the reporter on the right side.
RESULTS AND DISCUSSION
In the present work we have described a Taqman multiplex PCR assay for the quantitative analysis of the gene amplification phenomenon in cancer that allows both highly sensitive quantitative measurement of the relative number of oncogene copies in cancer cell lines.
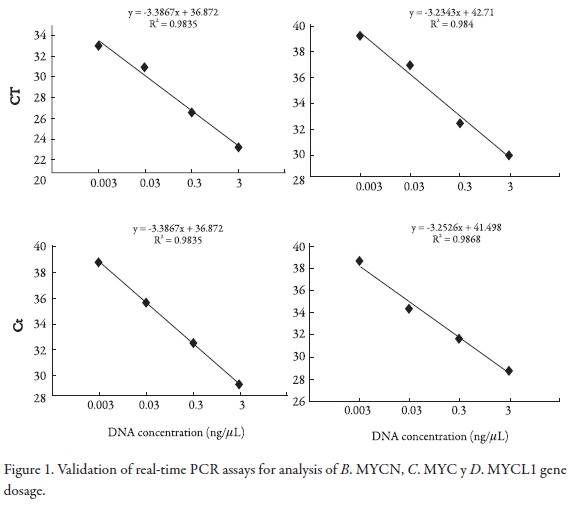
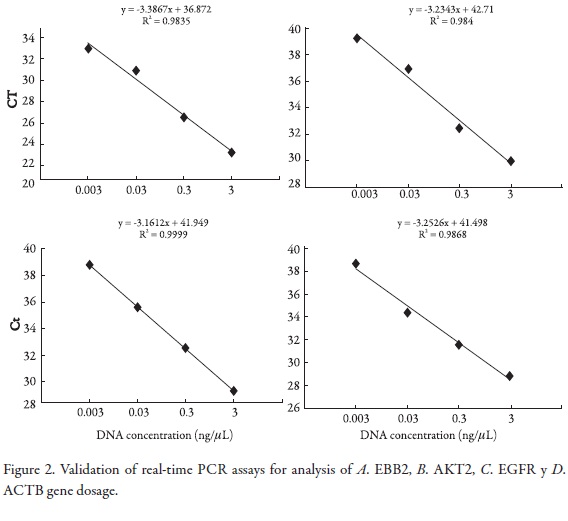
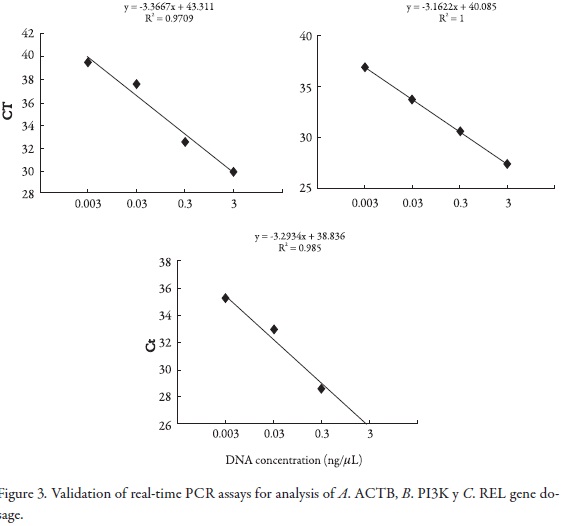
First, we demonstrated the feasibility of applying the Taqman multiplex assay by optimizing the gene dosage assays. For this, validation experiments were performed to determine the amplification efficiencies of primers and probes of oncogenes reported in the Cancer genome project database: EGFR, ERBB2, AKT2, MYC, MYCN, MYCL1, PI3KCA and REL with DNA serial dilutions within multiplex PCR. The efficiency values were measured using the CT slope method; this method involves generating a dilution series of the target template and determining the CT value for each dilution. A plot of CT versus log DNA concentration was constructed for each gene in the multiplex PCR. Efficiency calculation from the slopes of the calibration curve according to the equation E = 10 [–1/slope] (15, 16, 17). The slopes of the trend lines were (-3.386), (-3.234), (-3.161), (-3,252) for ACTB, MYCN, MYC and MYCL1 genes respectively, (Figure 1); (-3.207), (-3,396), (-3.39), (-3.286) for ERBB2, AKT2, EGFR and ACTB respectively (Figure 2) , and (-3.266), (-3,162), (-3.293) for ACTB, PI3KCA and REL genes respectively (Figure 3).
Simultaneous quantitative real-time PCR was run for the target genes and A. ACTB (reference). using various amounts of template DNA per reaction. CT values were determined and plotted against log input DNA for both genes. The resulting best fit line equations were: y = -3.386x + 36.87, -3.234x + 42.71, -3.161x + 42.94 and y = -3.252x + 41.49 for genes, respectively. Best fit line slopes were approximately equal with high linear correlation (R2 = 0.983 and 0.984, 0.999 and 0.986). The amplification efficiency of the Taqman assays was 2.04, 1.97, 2.03 y 2.07 respectively.
Simultaneous quantitative real-time PCR was run for the target genes and ACTB (reference) using various amounts of template DNA per reaction. CT values were determined and plotted against log input DNA for both genes. The resulting best fit line equations were: y = -3.207x + 42.61, -3.396x + 38.8, -3.390x + 40.89 and y = -3.286x + 42.95 for the genes, respectively. Best fit line slopes were approximately equal with high linear correlation (R2 = 0.998 and 0.986, 0.995 and 0.974). The amplification efficiency of the Taqman assays was 2.05, 1.96, 1.97 and 2.01 respectively.
Simultaneous quantitative real-time PCR was run for the target genes and ACTB (reference) using various amounts of template DNA per reaction. CT values were determined and plotted against log input DNA for genes. The resulting best fit line equations were: y = -3.366x + 43.31, -3.162x + 40.08 and -3.293x + 38.83 and for genes, respectively. Best fit line slopes were approximately equal with high linear correlation (R2 = 0.97 and 1, and 0,985). The amplification efficiency of the Taqman assays was 1.9, 2.07 and 2.01 respectively.
The reliability of the PCR reaction efficiencies was also assessed by plotting ΔΔCT values (Ct target - Ct ACTB) against the amount of input DNA in log scale. Through a wide range of template DNA input (6-100 ng), the absolute value of the trend line slopes for all genes were = 0.1, which indicated the validity of the relative quantitative assay by Δ ΔCT method..
We assessed the relative numbers of oncogenes copies gene copy number in relation to the copy number of a reference gene B-actin which is assumed to be 1 per haploid genome (Multiplex TaqMan assay, relative quantification). Genomic DNA was used to adapt and validate the multiplex PCR to measure the relative number of EGFR, ERBB2, AKT2, MYC, MYCN, MYCL1, PI3KCA and REL copy number in the panel of cancer cell lines.
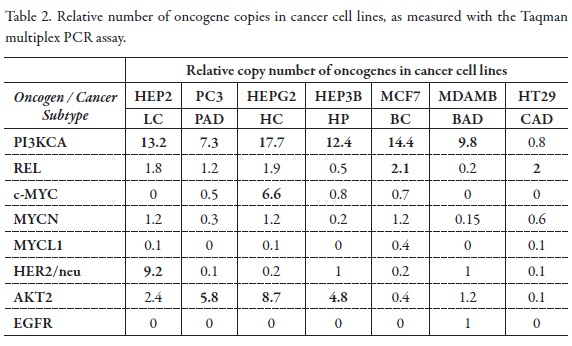
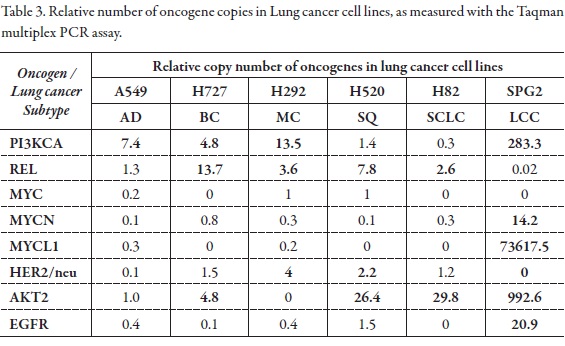
Change in PI3KCA gene dosage was observed in 76.9% of cell lines, AKT2 and REL in 46.15%; ERBB2 in 23%; MYC, MYCN, MYCL1 and EGFR in 7.69%. With this method the cells exhibited a gene dosage of REL from 2 to 13 times; ERBB2 gene from 2 to 9 times; MYC from 2 to 6 times; MYCN gene dose from 2 to 14; MYCL1 from 2 to 73617,5; EGFR from 2 to 21 and AKT2 from 2 to 992 (Figure 4,Figure 5, Table 2, Table 3).
During carcinogenesis, genetic and epigenetic alterations drive the evolution of tumor towards increased malignancy and treatment resistance. The changes enable tumor cells to overcome microenvironmental constraints, sustain proliferation, and invade adjacent tissues and distinct organs (13). Gene dosage alterations like gains and losses regulate the expression of genes and are motive forces for this evolution (8, 14). Tumor cells bearing an increasing number of gains and losses successively emerge and are selected for based on the growth advantage caused by the genetic changes. Discovery and functional assessment of gene dosage alterations involved in carcinogenesis are therefore essential for understanding the biology of the disease.
The AKT2 gene is a serine/threonine kinase key intermediate of signaling pathways that regulate cellular processes controlling cell size/growth, proliferation, survival, glucose metabolism, genome stability, and neo-vascularization (2). The gene was shown to be amplified and overexpressed in 2 of 8 ovarian carcinoma cell lines and 2 of 15 primary ovarian tumors (8), and breast carcinomas (3). Overexpression contributes to the malignant phenotype of a subset of human pancreatic ductal cancers. In this study, the Hep2 cell lines showed an increase in AKT2 gene copy to 2,4; PC3 cell line of 5,8; HepG2 cell line of 8,7; Hep3B of 4,8: H727 4,8; H520 of 26,4; H82 of 29,8; and SPG2 cell line 992,6 (Table 2, Table 3).
The PI3K/Akt signaling network is crucial to widely divergent physiological processes that include cell cycle progression, differentiation, transcription, translation and apoptosis (4, 12). It has been reported that the activated PI3K/Akt pathway provides major survival signals to prostate and many other cancer cells (7). Inhibition of Akt activity stimulates apoptosis in a wide range of mammalian cells (24). Therefore, the PI3K/Akt pathway is an attractive target for the development of of novel therapeutic strategies in patients with various tumor types. In this study, PI3KCA gene gain copies in the cell lines HEP2 (13,2), PC3 (7,3), HEPG2 (17,7), HEP3B (12,4), MCF7 (14,4), MDAMB (9,8), A549 (7,4), H727 (4,8), H292 (13,5) and SPG2 (283,3).
While REL has mostly been implicated in lymphoid tumors, a small scale of human non-small cell lung carcinomas (NSCLC) revealed elevated REL expression (25). Thus, the association of c-rel with human cancers may not be limited to lymphoid cells and a comprehensive analysis of c-rel in solid tumors may be particularly informative. Rel was amplified in the cell line of breast cancer MCF7 (2,1) and the lung cancer cell lines H727 (13,7), H292 (3,6), H520 (7,8) and H82 (2,16).
EGFR is a 170 kDa receptor tyrosine kinase (RTK) belonging to the ErbB receptor family. Upon EGF binding, EGFR dimerizes and activates downstream intracellular signal cascades, including Ras / Raf, JAK-STAT and phosphatidyl inositol 3-kinase (PIK3CA) / Akt; this results in the modulation of genes involved in cell proliferation, metastasis, angiogenesis and inhibition of apoptosis (11, 27). Several groups have reported that high polysomies and EGFR gene amplification predict favorable treatment response to TKI correlating with increased survival rate. EGFR gene amplification leads to protein over-expression and has been correlated with poor prognosis (28). In this study only the cell line obtained from a Colombian lung cancer patient, SPG2, showed amplification of EGFR gene with 20.9 copies. EGFR gene copy number is a parameter frequently considered to personalize the treatment because it is frequently determined of the efficacy of some drugs used in chemotherapy (29).
The HER2/neu protooncogene is located on the long arm of chromosome 17 (17q21), encoding for a transmembranal glycoprotein (p185neu) with intrinsic tyrosine kinase activity and marked sequence homology with the epidermal growth factor receptor (EGFR). Dimerization of HER2/neu with an activated EGFR molecule leads to the activation of a signal transduction cascade with subsequent increase in cell proliferation, angiogenesis, and metastatic potential, as well as a decrease in apoptosis. Amplification and/or over-expression of this gene has been reported in numerous cancers, including breast and ovarian tumors. In this study amplification of Her2/neu gene was observed in the cell lines HEP2 (9.2 copies), H292 (4 copies) and H520 cell line with 2.2 copies. The cell lines from breast carcinoma MCF7 and MDAMB were no amplified in this study as reported by (17).
The human MYC family of proto-oncogenes is among the most studied genes in cancer (31). Amplification of MYCN has emerged as among the clearest genetic indicators of high-risk, aggressive disease (5, 33). MYC family members (c-MYC, MYC and MYCL1) show differential expression in normal tissues. Amplification of the proto-oncogene MYCN occurs in 25% of tumors and is the best characterized genetic-risk factor for high-risk chemotherapy-refractory disease (32, 33, and 34). In this study the gene dosage of c–MYC gene occurred in the hepatocellular carcinoma cell line HEPG2 with 6.6 copies, as reported in the literature the expression of the proto-oncogene c-myc has been implicated in liver regeneration and hepatocarcinogenesis and the amplification is an indicator of malignant potential and poor prognosis in hepatocellular carcinoma (18).
The MYCN and MYCL1 genes were amplified in the Colombian patients cell line SPG2 with 14, 2 and 73617,5 copies respectively. MYCN was originally identified by oncogene expression profiling of human neuroblastoma cells (36). These surveys quickly established that, with few exceptions, cultured neuroblastoma cell lines carry the gene MYCN in amplified form. At the same time, neuroblastoma tumors were also found to carry amplified MYCN. The initial surveys suggested that MYCN amplification was specific for neuroblastoma. It turned out later that MYCN amplification can be seen in small cell lung cancer, retinoblastoma, malignant gliomas, and peripheral neuroectodermal tumors, although at a much lower incidence. As a common feature, all these tumors have neural qualities. Amplification values usually range between 50- and 100-fold, although much higher values have been reported in some cases. There is little data about c-myc DNA amplification in histological types of lung cancer other than small cell carcinoma.
This method calculates the copy number of each oncogen and is a promising alternative technique to FISH and Southern blot. Therefore, this technique should be considered as a powerful method gene dosage quantitation in clinical and research genetic surveys.
AGRADECIMIENTOS
The authors thank the Department of Pharmacy, Pharmacogenetics Cancer Group, and Universidad Nacional de Colombia, to facilitate the development of this work.
REFERENCIAS
1. D.G. Albertson, Gene amplification in cancer, Trends Genet., 22, 447-455 (2006). [ Links ]
2. A. Bellacosa, D. de Feo, A.K. Godwin, DW. Bell, J.Q. Cheng, D.A. Altomare, M .Wan, L. Dubeau, G. Scambia, V. Masciullo et al., Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas, Int. J. Cancer, 64, 280-285 (1995). [ Links ]
3. A. Bellacosa, C.C. Kumar, A. Di Cristofano, J.R. Testa, Activation of AKT kinases in cancer: Implications for therapeutic targeting, Adv. Cancer Res., 94, 29-86 (2005). [ Links ]
4. D.P. Brazil, Z.Z. Yang, B.A. Hemmings, Advances in protein kinase B signalling: AKTion on multiple fronts, Trends Biochem. Sci., 29, 233-242 (2004). [ Links ]
5. G.M. Brodeur, R.C. Seeger, M. Schwab, H.E. Varmus, J.M. Bishop, Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage, Science, 224, 1121-1124 (1984). [ Links ]
6. H. Chen, Y. Lin, L. Zhongfang, Y. Jun, Z. Junjie, D. Xu, L. Liying, W. Quanli, S. Tusheng, Detection of CCND1 amplification using laser capture microdissection coupled with real-time polymerase chain reaction in human esophageal squamous cell carcinoma, Cancer Gen. Cytogen., 175, 19e25 (2007). [ Links ]
7. W.S. Chen, P.Z. Xu, K. Gottlob, M.L. Chen, K. Sokol, T. Shiyanova et al., Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene, Genes Dev., 15, 2203-2208 (2001). [ Links ]
8. J. Cheng, A. Godwin, A. Bellacosa, T. Taguchi, T. Franke, T. Hamilton, P. Tsichlis, J. Testa, AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas, Proc. Natl. Acad. Sci. USA, 89, 9267-9271 (1992). [ Links ]
9. P.W. Chiang, W.J. Song, K.Y. Wu, J.R. Korenberg, E.J. Fogel, M.L. Van Keuren, D. Lashkari, D.M. Kurnit, Use of a fluorescent-PCR reaction to detect genomic sequence copy number and transcriptional abundance, Genome Res., 6,1013-1026 (1996). [ Links ]
10. P.W. Chiang, W.L. Wei, K. Gibson, R. Bodmer, D.M. Kurnit, A fluorescent quantitative PCR approach to map gene deletions in the Drosophila genome, Genetics, 153, 1313-1316 (1999). [ Links ]
11. E. Forgacs, S. Zöchbauer-Müller, E. Oláh, J.D. Minna, Molecular genetic abnormalities in the pathogenesis of human lung cancer, Pathol. Oncol. Res., 7(1), 6-13 (2001). [ Links ]
12. M. Hanada, J. Feng, B.A. Hemmings, Structure, regulation and function of PKB/AKT-a major therapeutic target, Biochim. Biophys Acta, 1697, 3-16 (2004). [ Links ]
13. D. Hanahan, R.A. Weinberg, The hallmarks of cancer, Cell, 100, 57-70 (2000). [ Links ]
14. C.A. Heid, J. Stevens, K.J. Livak, P.M. Williams, Real time quantitative PCR, Genome Res., 6, 986-994 (1996). [ Links ]
15. R. Higuchi, C. Fockler, G. Dollinger, R. Watson, Kinetic PCR analysis: Real-time monitoring of DNA amplification reactions, Biotechnology, 11(9), 1026-1030 (1993). [ Links ]
16. P.M. Holland, R.D. Abramson, R. Watson, D.H. Gelfand, Detection of specific polymerase chain reaction product by utilizing the 5'-3' exonuclease activity of Thermus aquaticus DNA polymerase, Proc. Natl. Acad. Sci. USA, 88, 7276-7280 (1991). [ Links ]
17. E. Hyman, P. Kauraniemi, S. Hautaniemi, M. Wolf, S. Mousses, E. Rozenblum, M. Ringner, G. Sauter, O. Monni, A. Elkahloun, O.P. Kallioniemi, A. Kallioniemi, Impact of DNA amplification on gene expression patterns in breast cancer, Cancer Res., 62, 6240-6245 (2002). [ Links ]
18. S. Kawate, T. Fukusato, S. Ohwada, A. Watanuki, Y. Morishita, Amplification of c-myc in hepatocellular carcinoma: Correlation with clinicopathologic features, proliferative activity and p53 overexpression, Oncology, 57(2), 157-63 (1999). [ Links ]
19. J.L. Kenneth, D. Thomas, Analysis of relative gene expression data using real-time quantitative PCR and the 2DDCT method, Methods, 25, 402-408 (2001). [ Links ]
20. S. Knuutila, A.M. Bjorkqvist, K. Autio, M. Tarkkanen, M. Wolf, O. Monni, J. Szymanska, M.L. Larramendy, J. Tapper, H. Pere, W. El-Rifai, S. Hemmer, V.M. Wasenius, V. Vidgren, Y. Zhu, DNA copy number amplifications in human neoplasms: Review of comparative genomic hybridization studies, Am. J. Pathol., 152, 1107-1123 (1998). [ Links ]
21. S. Knuutila, Y. Aalto, K. Autio, A.M. Bjorkqvist, W. El-Rifai, H. Pere, J. Tapper, M. Tarkkanen, A. Varis, V.M. Wasenius, M. Wolf, Y. Zhu, DNA copy number losses in human neoplasms, Am. J. Pathol., 155, 683-694 (1999). [ Links ]
22. H. Lei, P.J. Furlong, J.H. Ra, D. Mullins, R. Cantor, D.L. Fraker, F.R. Spitz, AKT activation and response to interferon in human cancer cells, Cancer Biol. Ther., 4, 709-715 (2005). [ Links ]
23. N. Meyer, L.Z. Penn, Reflecting on 25 years with MYC, Nat. Rev. Cancer, 8, 976-990 (2008). [ Links ]
24. Y. Mosesson, G.B. Mills, Y. Yarden, Derailed endocytosis: An emerging feature of cancer, Nat. Rev. Cancer, 8, 835-850 (2008). [ Links ]
25. T. Mukhopadhyay, J.A. Roth and S.A. Maxwell, Altered expression of the p50 subunit of the NF-kappa B transcription factor complex in non-small cell lung carcinoma, Oncogene, 11, 999-1003 (1995). [ Links ]
26. N. Normanno, A. de Luca, C. Bianco, L. Strizzi, M. Mancino, M.R. Maiello, A. Carotenuto, G. de Feo, F. Caponigro, D.S. Salomon, Epidermal growth factor receptor (EGFR) signaling in cancer, Gene, 366(1), 2-16 (2006). [ Links ]
27. R. Rasmussen, In "Rapid cycle real-time PCR, methods and applications: Quantification on the cight cycler". Edited by S. Meuer, C. Wittwer, K. Nakagawara, Springer Press, Heidelberg (2001). [ Links ]
28. R. Redon, S. Ishikawa, K.R. Fitch, L. Feuk, G.H. Perry, T.D. Andrews, H. Fiegler, M.H. Shapero, A.R. Carso, W. Chen et al., Global variation in copy number in the human genome, Nature, 444(7118), 444-454 (2006). [ Links ]
29. R.D. Riley, D. Heney, D.R. Jones, A.J. Sutton, P.C. Lambert, K.R. Abrams et al., A systematic review of molecular and biological tumor markers in neuroblastoma, Clin. Cancer Res., 10, 4-12 (2004). [ Links ]
30. A. Sakurada, M.S. Tsao, Predictive biomarkers for EGFR therapy, IDRUGS, 12, 34-38 (2009). [ Links ]
31. M. Schwab, K. Alitalo, K.H. Klempnauer, H.E. Varmus, J.M. Bishop, F. Gilbert, G. Brodeur, M. Goldstein, J. Trent, Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumor, Nature, 305, 245-248 (1983). [ Links ]
32. R.C. Seeger, G.M. Brodeur, H. Sather, A. Dalton, S.E. Siegel, K.Y. Wong et al., Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas, N. Engl. J. Med., 313, 1111-1116 (1985). [ Links ]
33. G. Selvaggi, S. Novello, V. Torri, E. Leonardo, P. de Giuli, P. Borasio, C. Mossetti, F. Ardissone, P. Lausi, G.V. Scagliotti, Epidermal growth factor receptor overexpression correlates with poor prognosis in completely resected non-small-cell lung cancer, Ann. Oncol., 15(1), 28-32 (2004). [ Links ]