INTRODUCTION
Advances in technology have made multigene testing, or "panel testing", a practical option when looking for genetic variants that may be associated with a risk of breast cancer and treatment outcome. Use of pharmacogenomics will optimize treatment and lower health costs. In this context, it is necessary to study disease associations and pharmacogenetic polymorphisms in populations to optimize disease treatment. In the case of breast cancer, tamoxifen metabolism related polymorphisms are associated to treatment, but their frequency is poorly known in Colombian population.
Using different platforms to process and analyze samples can simultaneously identify genetic variants associated with susceptibility, clinical outcomes and drug response. For example, the spectrum of the frequency of the MTHFR gene polymorphism C677T in a group of healthy Colombian individuals was previously determined to evaluate the genetic background in relation to disease susceptibility and pharmacogenetic applications; the results for the C/C and C/T genotypes in a Colombian population were similar to other previously studied groups of healthy subjects. Subjects from Colombian populations might be at risk of developing diseases associated with MTHFR polymorphisms and might present toxicity and adverse effects if treated with MTX, which suggests the need to evaluate therapeutic alternatives based on individual pharmacogenetic studies [1].
In the case of breast cancer, the response to several hormone therapeutics, such as tamoxifen are mediated by the action of CYP2D6, which is highly polymorphic, and genotypes has been identified allowing classifying patients as ultra-rapid, normal and poor metabolizers.
Genetic variants in certain genes have been associated with early cancer risk [2], risk of developing cancer [3] and susceptibility to cancer [4]. In this way, CYP2D6*4 variant (rs3892097) is associated with response to tamoxifen, since patients carrying a T allele will present slow metabolism of this drug and would therefore need a dose adjustment or an alternative therapy [5]. In the case of CAS8 rs1045485, its C allele is associated with a lower risk of breast cancer [6,7]. On the other hand, ESR1 rs2046210 has been associated with risk of developing breast and endometrial cancer in Chinese women [8-10], and variant C has been associated with ER-negative but not ER-positive breast cancer (P > 0.05). These findings provide further evidence for distinct etiological pathways associated with invasive ER-positive and ER-negative breast cancer in younger women and women of African ancestry [11]. FGFR2rs1219648 showed a similar association with the risk of developing breast cancer in Chinese [12] and Indian populations [13]. Polymorphism rs13387042-2q35(G/A), has been associated with breast cancer risk only among postmenopausal women who never used hormone therapy [14]. Furthermore, the GG genotype of SNP rs13281615 plays a role in breast cancer through PVT1 expression during oncogenesis, and "protective" mutations could occur [15]. MSRP30 rs10941679 was significantly associated with BCIS (Breast cancer in situ) risk [16]. As for TNRC9 rs3803662, allele GG increased the risk of breast cancer in codominant inheritance (OR=2.19, 95% CI:1.19-4.02), and recessive genetic models (OR=2.06, 95% CI: 1.15-3.70) [17]. Moreover, the variant rs6964587 was associated with increased breast edema due to radiotherapy after the following 5 years (Beta, 0.22; 95% confidence interval, 0.09-0.34; P = 7 x 10-4) [18] and increased to risk of breast cancer [19]. LSP1 rs3817198 has been associated with mammographic density in genome-wide studies [20]. Likewise, MAP3K1rs889312, has been associated with breast cancer risk in women of European ancestry [21] and ESR1rs3020314 showed stronger associations in ER-positive [22]. Finally, in AKAP9, the minor T allele of 7q21-rs6964587 was associated with breast cancer risk under a recessive model (OR 1.07, 95% CI 1.00 to 1.13, p = 0.04) [23].
Knowledge of Colombian disease associated polymorphism frequencies will allow to establish National Health policies to improve health and treatment outcome.
METHODS
Population
A retrospective cross-sectional study in 160 healthy Colombian individuals over 18 years was conducted using molecular genetic profiling, age ranges between 18 and 74 years with a mean of 40.07 years. DNA samples were collected from saliva samples in Instituto de Referencia Andino, as well as all corresponding applications from medical orders and/or individuals received between the years 2012 and 2014. All individuals signed an informed consent.
The Healthy Woman DNA Insight™ panel from Pathway Genomics® evaluates panel of breast cancer (CAS8 rs1045485, CHEK21100delC, ESR1 rs2046210, FGFR2rs1219648, intergenic_2q35rs13387042, intergenic_8q24 rs13281615, MSRP30 rs10941679, TNRC9 rs3803662, AKAP9 rs6964587, LSP1 rs3817198, MAP3K1rs889312, PALBS1592 delT, ESR1rs3020314). The pharmacogenetics (CYP2D6*4), in addition to Abacavir® hypersensitivity, aminoglycoside-induced ototoxicity, response to beta blockers, carbamazepine hypersensitivity, clopidogrel metabolism, estrogen supplementation, alpha interferon, metoprolol metabolism, metabolism and hypersensitivity to phenytoin, proton-pump inhibitor, simvastatin induced myopathy, metabolism of voriconazole and warfarin.
DNA microarrays were used to measure expression levels of probes, corresponding to alleles in CAS8 rs1045485, CHEK21100delC, ESR1 rs2046210,FGFR2rs1219648, intergenic_2q35rs13387042, intergenic_8q24 rs13281615, MSRP30 rs10941679, TNRC9 rs3803662,AKAP9 rs6964587, LSP1 rs3817198, MAP3K1rs889312, PALBS1592 delT, ESR1rs3020314 genes. Each microarray position contains 10-12 picomoles of a specific DNA sequence under high stringency. Probe-target hybridization was detected and quantified by fluorophores attached to the probe.
The frequency of the polymorphism substitution in the genes markers was analyzed. Each participant was identified by a code used in a database, where the descriptive analysis for continuous variables such as age was performed with measures of central tendency and Shapiro-Wilk normality test. The description of categorical variables (origin, sex, genotype and allele) was performed by frequency analysis. Given the variable nature of the study association levels, they were evaluated by a Chi square test (X2) with a confidence level of 95%. In addition, Hardy-Weinberg equilibrium was tested. Data base designed in Access includes the following variables that were Collected: Code anonymity of each individual, age, gender, date of birth, maternal ancestry, paternal ancestry, nationality, CYP2D6 rs3892097, panel for breast cancer (CAS8 rs1045485, CHEK21100delC, ESR1 rs2046210, FGFR2rs1219648, intergenic_2q35rs13387042, intergenic_8q24 rs13281615, MSRP30 rs10941679, TNRC9 rs3803662,AKAP9 rs6964587, LSP1 rs3817198, MAP3K1rs889312, PALBS1592 delT, ESR1rs3020314).
Statistical Methods
Available results were tabulated to perform the respective calculation of frequencies of each polymorphism and Hardy-Weinberg equilibrium was calculated using the online Curt Lab - HW calculator, and then a Genpop software was used in order to define genetic frequencies. Later on, contingency tables were established with a double entry and Chi squared distribution calculated. Results were transferred to the SPSS statistical program for Windows V20. They were not stratified by ethnic group or socioeconomic levels. Available frequencies were collected in HapMap Genome Browser release #28 (Phases 1, 2 and 3 - merged genotypes and frequencies) for the general population and also for the Colombian population for each of the variants tested in this review.
RESULTS
No deviation from Hardy-Weinberg equilibrium is present; therefore, the population included in this study showed no significant migration, subgroup stratification, inbreeding or selection processes. Additionally, by contrasting data reported in Hap-Map for general population, Colombians of the present study have no significant differences with the sample of Colombian population reported in HapMap.
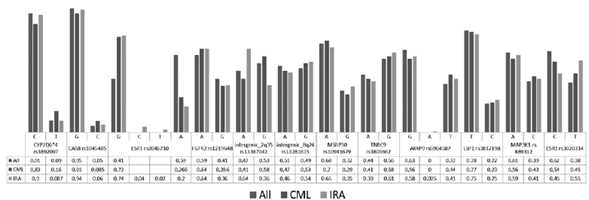
After evaluatingeach particular polymorphism, CYP2D6*4 rs3892097, CAS8rs1045485, MSRP30 rs10941679, LSP1 rs3817198 and MAP3K1 rs889312, data found in the present study are similar to data report for general and Colombian population previously evaluated in HapMap. Allele Frequency Comparison between Present Study and Hap-Map is shown in Table 1.
Table 1 Allele frequency comparison between a Colombian population in the present study and HapMap database.
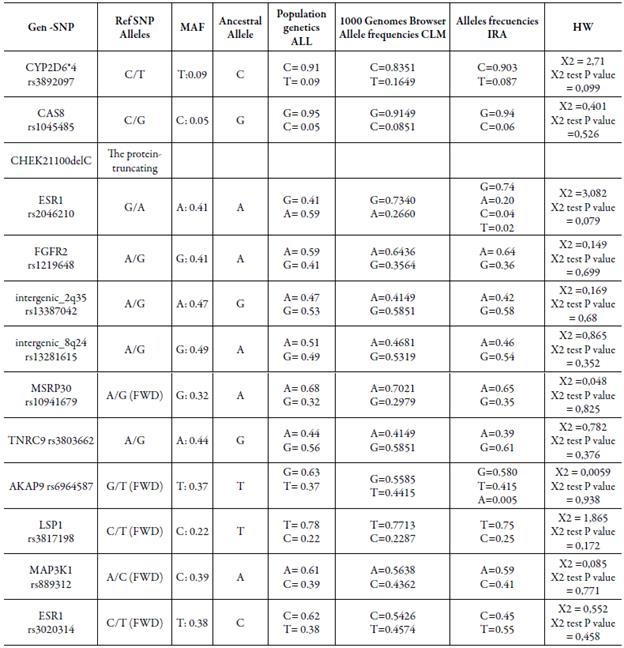
Polymorphisms FGFR2 rs1219648, intergenic 2q35-rs13387042, intergenic 8q24-rs13281615, TNRC9 rs3803662 and ESR1 rs3020314, showed higher frequencies than those reported previously for the Colombian population in HapMap, and has greater similarity to those reported for the global population.
In addition, we identified in ESR1 rs2046210 variants C and G have frequencies of 0.04 and 0.02, respectively; similarly, AKAP9 rs6964587 allele A was found in a frequency of0.005. The presence of these variants had not been previously reported in the Colombian population or the HapMap database.
DISCUSSION
As we move from gene disease discovery into the application of genetic knowledge, allele frequency studies in populations become more important for the design, implementation of studies and for determining treatment according to personal polymorphisms. Studies in Colombian population are scarce, and the present study determined the allele frequencies of 14 polymorphisms, previously associated with breast cancer status. Allele frequencies were compared with dbSNP and other data sources in HapMap.
Allele frequencies for minor alleles relevant for Colombian population are here reported, although the number of individuals within the categories is small. Local differences in allele frequencies should be considered when designing association studies in the future. One weakness of our analysis is the lack of treatment follow-up due to the diversity of places where patients are treated.
The number of useful biomarkers for predicting drug response will rapidly increase for improving treatment and settle the basis for personalized medicine. Pharmacogenetic profiles will be useful for prevention programs, and personalized therapies will reduce health costs and ensure a more efficient health care.
Conclusion
The population studied was not significantly different in allele distribution with previously reported data from HapMap. Genotypes in Colombian population are similar to other previously studied groups of healthy subjects. The use of genotyping pharmacogenetic polymorphisms will prevent toxicity and adverse effects in tamoxifen treatment (for example in CYP2D6 rs3892097), and therapeutic alternatives should be evaluated based on individual pharmacogenetic studies.