INTRODUCTION
Escherichia coli is a Gram-negative bacillus that belongs to the Enterobacteriales order and is part of the intestinal microbiota of humans and animals. It is also considered an ubiquitous pathogen, which can be found in several environments, such as rivers, lakes, sewage, and soil. Furthermore, E. coli is also one of the most commonly isolated species from clinical samples, in which it causes several diseases, including pneumonia, abscesses, septicemia, otitis, gastrointestinal infections, and urinary tract infections [1].
E. coli infections can be treated with several classes of antimicrobials. However, due to an alarming increase of E. coli with diversified antimicrobial resistance phenotypes, the number of therapeutic options has decreased considerably in recent years [2].
Aminoglycosides are antimicrobial agents obtained from the secondary metabolism of bacteria of the Streptomyces and Micromonospora genus. Neomycin, tobramycin, kan-amycin, gentamicin, and amikacin are the main representatives of this pharmacological class, which is available for clinical use since the 1943s [3]. The most recent drug developed of this class is plazomycin, approved in 2018 by the FDA and obtained by structural modifications of existing aminoglycosides due to the need to circumvent antimicrobial resistance [4]. Aminoglycosides are concentration-dependent bactericide agents that act through inhibition of protein synthesis and have a broad spectrum of action with increased activity against Enterobacteriales [4, 5]. These antimicrobials are available for clinical administration via inhalational, topical, oral, intramuscular, and intravenous formulations for the empirical therapy of serious infections caused by mul-tidrug-resistant pathogens [4, 6].
Despite the potent bactericidal effect of aminoglycosides, their indiscriminate use has selected many resistant microorganisms, especially E. coli. Bacterial resistance against aminoglycosides can be chromosomal or encoded by mobile genetic elements, such as plasmids and transposons. Currently, four aminoglycoside-resistance mechanisms are known, which include (i) enzymatic modification and inactivation of the drug, (ii) increased efflux of the antimicrobial, (iii) decreased membrane permeability, and (iv) modifications of the pharmacological target [4, 5].
Enzymatic modification/inactivation is the most frequent resistance mechanism to aminoglycosides in Enterobacteriales and involves aminoglycoside-modifying enzymes (AME) that are classified into three groups: O-phosphotransferase (APH), amino-glycoside O-nucleotidyltransferase (ANT), and aminoglycoside N-acetyltransferase (AAC). These enzymes are encoded by genes frequently carried by plasmids and transposons with pronounced potential for interspecies dissemination [7-9]. Furthermore, the report of mobile genetic elements harboring AMEs and determinants of resistance to beta-lactam antimicrobials, such as extended-spectrum beta-lactamase enzymes (ESBL) and carbapenemases, is of particular concern since they can induce multidrug resistance (MDR) or extensively drug-resistant (XDR) phenotypes in Gram-negative bacteria [10, 11]. ESBL-positive bacterial isolates are resistant to penicillins, cephalosporins, and monobactams, while carbapenemases provide resistance to carbapenems, considered the latest generation of beta-lactams. In this context, E. coli AMEs- and ESBL-positive have been reported in clinical isolates [12, 13] and chicken farms [14]. Importantly, in Brazil, this finding has extended beyond the clinical setting, in which these isolates were found in water samples [15], feedlot lambs [16], and peri-urban wild animals [17].
The detection of AMEs and ESBLs in bacterial isolates, especially in medical settings, guides the antimicrobial choice and avoids therapeutic failure. However, due to its diversity, AMEs detection must be done in the laboratory using a molecular approach, which is not yet a reality in the diagnosis routine. Thus, the search for inferences based on phenotypes has been developed [18, 19]. Therefore, considering the relevance of aminoglycosides in the clinical setting, we aimed to evaluate the resistance of clinical (CI) and domestic sewage (DS) E. coli isolates to aminoglycosides. In addition, the possible presence of AMEs was identified through phenotypic methods.
MATERIAL AND METHODS
Bacterial isolates
CI (n = 52, only urinary tract) and DS (n = 33) E. coli isolates used in this study were obtained in Divinópolis (MG) (geographical coordinates: 20° 08' 20'' S and 44° 53' 02'' W), located in the southeast of Brazil. DS isolates were characterized by Alves-Coelho et al. (2021) [20], while CI isolates belong to the collection of the Laboratório de Diagnóstico Laboratorial e Microbiologia Clínica/Universidade Federal de São João del-Rei, and the species identification was previously performed using a VITEK-2* automated system (bioMérieux, Marcy-I'Étoile, France). The isolates showed different resistance profiles to beta-lactams, including ESBL-producers (five DS and 22 CI isolates) previously determined using the approximation disk test according to the Clinical Laboratory Standard Institute (2017) [21]. All isolates were maintained as frozen stocks at -20 °C with 15% glycerol (Isofar, Brazil).
Aminoglycoside susceptibility testing
The minimal inhibitory concentration (MIC) of the aminoglycosides (Cecon, Brazil) amikacin (AMI), gentamicin (GEN), and neomycin (NEO) was determined by the microdilution method, following the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2021) [22].
E. coli isolates with a MIC > 8 µg/mL and > 2 µg/mL to amikacin and gentamicin, respectively, were classified as resistant. For neomycin, isolates with MIC < 8µg/mL were considered wild-type (WT) (without antimicrobial resistance mechanism) based on the epidemiological cut-off point (ECOFF). According to the EUCAST (2021) [22], the isolates cannot be categorized as resistant or sensitive to neomycin due to their lack of cut-off point. Therefore, the analysis of the WT phenotype of the iso lates is recommended (i.e., they do not present mechanisms of resistance to neomycin). Despite that, at some points in this work, we consider the absence of resistance mechanism as "sensitive" to facilitate the understanding of the context of neomycin susceptibility. E. coli American Type Culture Collection (ATCC) 25922 was used as a standard strain in antimicrobial susceptibility testing.
MIC50 and MIC90 values for gentamicin and amikacin were defined as the lowest concentrations that inhibited 50% and 90% of bacterial isolates, respectively.
Inferences of AME production from the aminoglycoside resistance phenotype
The disk diffusion test was performed to evaluate the presence of modifying enzymes in E. coli isolates as a possible aminoglycoside resistance mechanism [22].
The antimicrobials (Cecon, Brazil) amikacin (30 µg), gentamicin (10 µg), kanamycin (30 µg, KAN), and tobramycin (10 µg, TOB) were used. The results were analyzed and interpreted according to Mancini et al. (2019) [19], which proposed an algorithm to correlate the resistance phenotype with the production of AMEs by bacterial isolates.
RESULTS AND DISCUSSION
Aminoglycoside susceptibility testing
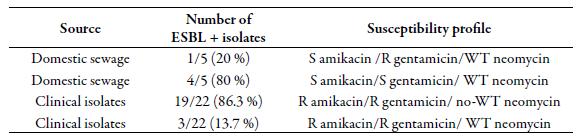
In this study, two distinct populations of E. coli were evaluated for their susceptibility profile to amikacin, gentamicin, and neomycin. The MICs and MIC50/MIC90 obtained revealed a significant difference between the susceptibility profiles of the isolates according to their origin. In general, 33.3% (11/33) of the DS (table 1) and 100% (52) of the CI (table 2) isolates showed mechanisms of resistance to amikacin, gentamicin, or neomycin. It is noteworthy that 78.8 % (41/52) of the CI isolates were resistant to the three aminoglycosides tested, while 69.9 % (23/33) of the DS isolates were sensitive to these drugs (table 2). Similarly, Lindemann et al. (2012) [23] showed that resistance to more than 3 aminoglycosides was frequent between clinical isolates of E. coli (mostly from urine) recovered from Western Norway, which corresponds to 92 % of these isolates. In another study, 66.19 % of 276 uropathogenic E. coli (UPEC) obtained from urine samples in a hospital in Tehran were resistant to gentamicin, confirming the high rate of aminoglycoside resistance among UPECs recovered from hospitalized patients [24].
Table 1 Minimum inhibitory concentration (MIC) values for aminoglycosides (amikacin, gentamycin, and neomycin) among Escherichia coli recovered from domestic sewage and phenotypic determination of mechanisms of resistance to aminoglycosides and beta-lactam.
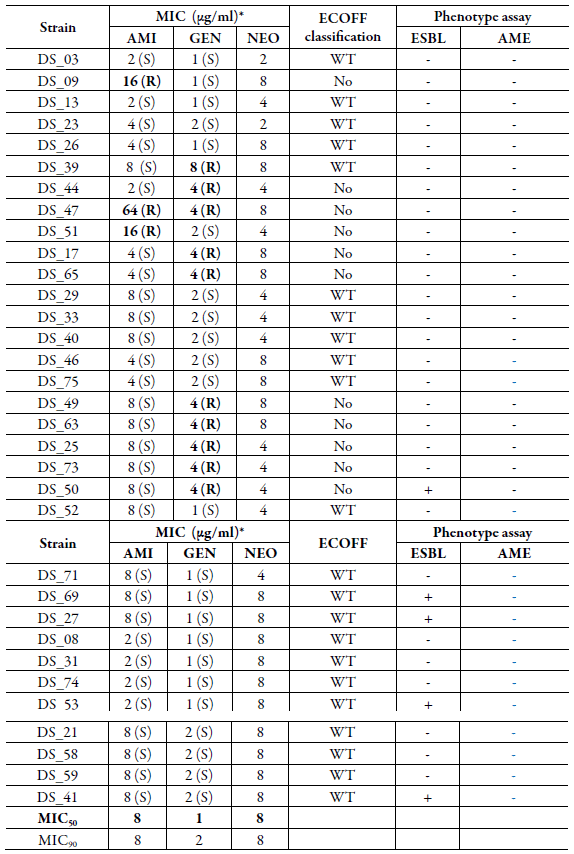
WT: Wild type, without antimicrobial resistance mechanism based on the epidemiological cutoff point (ECOFF - EUCAST 2021); No: Isolate no wild type; AMI: Amikacin; GEN: Gentamycin; NEO: Neomycin; R: Resistant; S: Susceptible; ESBL: Extended-spectrum beta-lactamase enzymes; AME: Aminoglycoside-modifying enzymes.
*According to EUCAST (2021), MIC > 8µg/mL and > 2µg/mL to amikacin and gentamicin, respectively, were categorized as resistant and MIC ≤ 8µg/mL for neomycin were considered wild type.
Table 2 Minimum inhibitory concentration (MIC) values for aminoglycosides (amikacin, gentamycin, and neomycin) among Escherichia coli recovered from urine sample of hospitalized patients with diagnostic of urinary infection and phenotypic determination of mechanisms of resistance to aminoglycosides and beta-lactam.
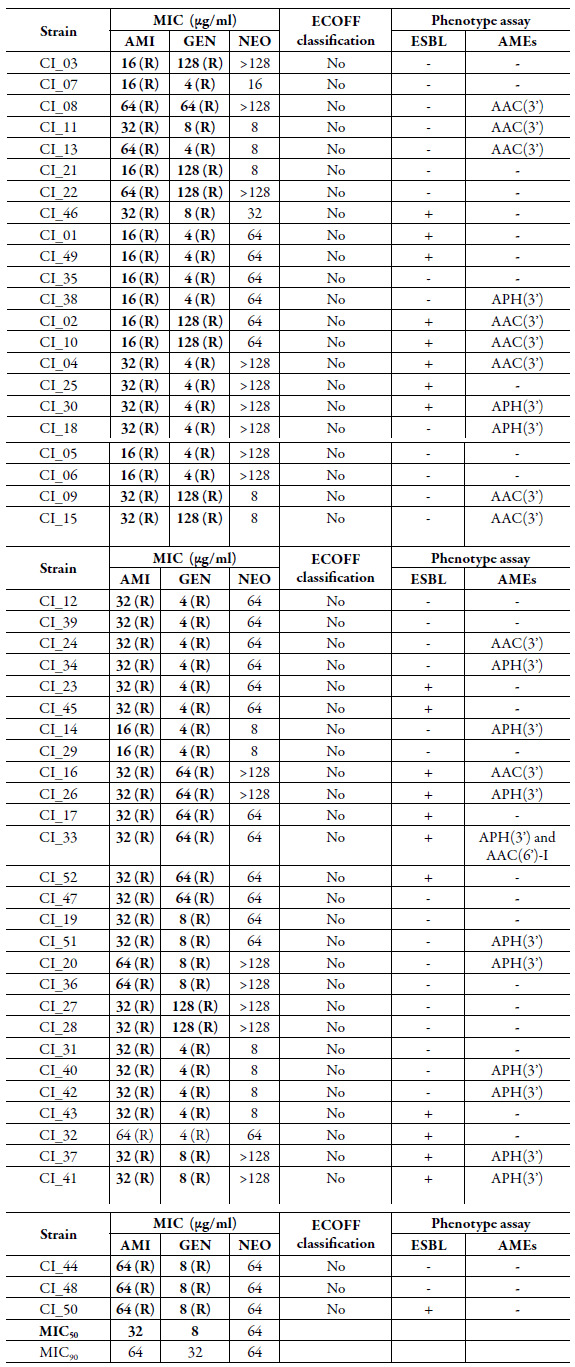
WT: Wild type, without antimicrobial resistance mechanism based on the epidemiological cutoff point (ECOFF -EUCAST 2021); No: Isolate no wild type; AMI: Amikacin; GEN: Gentamycin; NEO: Neomycin; R: Resistant; S: Susceptible; ESBL: Extended-spectrum beta-lactamase enzymes; AME: Aminoglycoside-modifying enzymes; AAC: Aminoglycoside acetyltransferases; APH: Aminoglycoside phosphotransferases.
*According to EUCAST (2021), MIC > 8µg/mL and > 2µg/mL to amikacin and gentamicin, respectively, were categorized as resistant and MIC < 8µg/mL for neomycin were considered wild type.
Considering the studied population (n = 82), 67.0 % (55) and 74.4 % (61) of the E. coli isolates were resistant to amikacin and gentamicin, respectively. Furthermore, resistance to amikacin and gentamicin occurred simultaneously in all CI isolates (table 2), while in DS isolates, this effect was only observed in the DS_47 (table 1). In contrast, Lindemann et al. (2012) [23] showed that amikacin resistance was lower in 67 clinical isolates of E. coli that exhibited resistance to aminoglycosides and extended-spectrum beta-lactam antibiotics. Still, a diversified profile was observed among the DS isolates, with 24.4 % (8/33) resistant to gentamicin and sensitive to amikacin and 6.0 % (2/33) resistant to only amikacin (table 1).
Regarding neomycin, all DS isolates were considered WT or without resistance mechanism, according to the ECOFF value (EUCAST, 2021) [22] (table 1). However, only 11 out of 52 clinical E.coli showed this phenotype, in which 17 isolates showed MICs higher than 128 µg/mL (table 2). These results reveal a significantly higher rate of neomycin resistance in Brazil compared to Europe. Indeed, low frequencies of resistance to this aminoglycoside were found in E. coli isolates in a retrospective study in Germany, in which the percentage of neomycin-resistant isolates decreased from 2006 to 2017 (18.4 % to 17.9 %) [25]. The difference between the two countries alludes to the indiscriminate use of neomycin-based ointments in Brazil compared to Europe [26-28], making the bacterial population more sensitive to selective pressure in South American countries.
ESBL production in Enterobacteriales has been described to occur in conjunction with AMEs, considering the common genetic elements involved in gene transfer [13]. In this context, the findings of this study again show different profiles among the populations of E. coli studied. It was observed that among ESBL-producing E. coli, four susceptibility profiles to amikacin, gentamicin, and neomycin were detected (table 3), differing based on their origin. The ESBL-positive DS isolates showed greater sensitivity to the aminoglycosides tested. In contrast, all CI isolates were resistant to amikacin and gentamicin, with MICs ranging from 16 - 64 µg/mL and 4 - 128 µg/mL, respectively. Finally, most of the ESBL-producing isolates studied here (23/27, 85 %) showed mechanisms of resistance to gentamicin and/or neomycin, in which simultaneous resistance to amikacin was observed only in clinical isolates.
Inferences of AME production from the aminoglycoside resistance phenotype
Aminoglycosides, especially amikacin and gentamicin, are antimicrobial agents of great relevance in the treatment of infections caused by multidrug-resistant Gram-negative bacteria. In these cases, they are usually administered in combination with other classes of antimicrobials, such as beta-lactams and quinolones [29, 30]. However, the widespread use of aminoglycosides favored the development of bacterial resistance, in which the main resistance mechanisms involve the action of AMEs or methyltransferases enzymes that modify the binding target. Among the AMEs extensively produced by E. coli, AAC, APH, and ANT often act on more than one type of substrate within the aminoglycoside class [9].
In this context, reference protocols do not provide tests capable of detecting the aminoglycoside resistance phenotype of bacteria, requiring the determination of AME-positive isolates by molecular approach. Considering the limited reach of this tool in microbiology laboratories, the search for alternatives is essential and can help to understand the susceptibility profile to aminoglycosides with inference in the therapeutic outcome. For instance, Mancini et al. (2019) [19] proposed the use of an algorithm based on the diameter of the halo of inhibition of aminoglycoside substrates (i.e., gentamicin, amikacin, tobramycin, and kanamycin), which, in comparison to the genotypic determination, was able to evaluate the production of some AMEs in E. coli.
Using this algorithm, we observed a diversity of AME-production profiles among the E. coli isolates studied. As shown in table 1, all DS isolates were considered WT-no AME (WT without AME resistance mechanism, according to Mancini et al. (2019) [19]). Among CI isolates, 55.7 % (29/52) presented this phenotype (table 2). It should be noted that all clinical isolates were recovered from patients with urinary tract infection, which suggests that these isolates are of the UPEC pathotype. Interestingly, this pathotype can also be found in other environments, such as domestic sewage, but apparently it was not present in the DS population studied. As reviewed by Jang et al. (2017) [2]), E. coli from natural environments seems to present particularities regarding the dissemination of antibiotic resistance and virulence that remains to be clarified.
APH (3') (14/52) and AAC (3') (10/52) enzymes were detected among CI isolates in this study. Additionally, one of the isolates (CI_33) was categorized as a producer of APH (3') and AAC (6')-I enzymes simultaneously (table 1). In fact, these enzymes are widely disseminated among Gram-negative bacteria, which can be attributed to their high potential for mobility via plasmids, transposons, and integrons [5].
APH (3')-positive isolates may have decreased susceptibility to neomycin, but gentamicin and amikacin are not substrates for this enzyme. In contrast, the production of AAC (3') and or AAC (6')-I compromises the susceptibility of Gram-negative bacteria to gentamicin and amikacin without interfering with that of neomycin.
Here, 76.9 % (10/13) and 60,0% (6/10) of APH (3')- and AAC (3')-positive CI isolates, respectively, had MIC ≥ 64 µg/mL for neomycin, indicating that these isolates are not WT according to EUCAST 2021 [22]. However, the finding of non-AME-producing isolates but with MIC ≥ 8 µg/mL for neomycin (WT no-AME based on the epidemiological cut-off point-ECOFF) indicates the presence of other mechanisms of resistance to this compound. It should be monitored since neomycin is widely used in the treatment of skin infections.
The co-occurrence of resistance to aminoglycosides and beta-lactam antimicrobials has been reported. Specifically, in isolates resistant to beta-lactams due to an enzymatic mechanism, including ESBL type, Bodendoerfer et al. (2020) [13] demonstrated that more than 50 % of the clinical isolates are resistant to amikacin or gentamicin, most often by the production of APH (3'), AAC (3') and AAC (6 '). Similarly, regarding the E. coli isolates included in this study, 40.7 % produce ESBL and AME simultaneously (table 2). Furthermore, resistance mechanisms to aminoglycosides and beta-lactams were observed exclusively in isolates of clinical origin, suggesting the impact of selective pressure of antimicrobials and the occurrence of horizontal transfer of genes for resistance to antimicrobials.
CONCLUSION
In conclusion, this study showed that resistance to aminoglycosides of clinical relevance is present in E. coli isolates in Brazil of both clinical and environmental origin, in the latter to a lesser extent. Furthermore, the results show that AMEs are frequent in clinical isolates with an imminent risk of dissemination and correlation with clinical therapeutic failure. Thus, epidemiological and molecular surveillance for aminoglyco-side resistance must be implemented to ensure better monitoring of cases of infections caused by E. coli resistant to this important class of antibiotics.