Introduction
Cancer is a disease characterized by the loss of cell cycle control and is a major cause of morbidity and mortality worldwide in its metastatic form.1-3 Traditionally, its management is based on three classic pillars: surgery, radiation therapy, and chemotherapy; these three alternatives, used alone or in combination, reduce tumor burden and stop the growth of malignant cells.4,5 Other strategies include targeted or hormone therapy and the use of inhibitors that interfere with the molecular mechanisms responsible for uncontrolled growth and the characteristic survival of cancer cells.6
In recent years, translational cancer research has made significant progress in its diagnosis and treatment. In addition, this type of research has contributed significantly to the positioning of immunotherapy as a fourth pillar for the treatment of this disease.7
Regarding cancer treatment, immunotherapy may be classified as passive or active.8 On the one hand, the administration of cellular and molecular components with antitumor activity, such as monoclonal antibodies, and adoptive cell transfer (ACT) are forms of passive immunotherapy; on the other hand, vaccines and antibodies that block inhibitory signals of T-cell activation (immune checkpoint inhibitors) are active immunotherapies.
Furthermore, the study of cancer immunobiology has shown that genomic instability and immune surveillance of tumors induce permanent changes in the tumor (immunoediting) that determine its uniqueness. Consequently, rational cancer management should include biomarker monitoring (for timely diagnosis) combined with the application of personalized treatment regimens based on the specific characteristics of each tumor.
This personalized treatment should begin with tumor preservation (which is mostly done only to confirm the histopathologic diagnosis) in order to: i) analyze the composition of the immune cell infiltrate to predict its evolution and the prognosis of clinical response;9-11 ii) monitor the sensitivity of tumor cells to different drugs in vitro to select those to be used in the patient;2-15 and iii) design different types of therapeutic vaccines using antigenic determinants of mutated proteins, lysates, or tumor ribonucleic acid (RNA) as a source of tumor antigen and dendritic cells (DC) of the patient, which are used as immunoadjuvant therapy and delivery vehicle of the antigen to the immune system.16-19
Vaccines whose design relies on the use of synthetic antigens based on somatic mutations expressed specifically by the tumor and not by normal cells (neoantigens) are a novel and efficient strategy for inducing antitumor T cells, since the T cells that recognize them have not experienced the induction of self-tolerance.
Given this scenario, it can be assumed that the use of personalized therapeutic neoantigen cancer vaccines will be a therapeutic alternative for this disease in the medium term; therefore, the aim of this review is to briefly describe the biological rationale, the current development prospects, and some clinical outcomes of personalized neoantigen-based cancer immunotherapy.
Materials and methods
A literature search was performed in PubMed, Scopus and EBSCO using the following search strategy: Type of articles: original experimental studies, clinical trials, and systematic narrative reviews on identification methods of cancer-associated mutations or cancer immunotherapy strategies with neoantigen-based vaccines generated by tumor genomic instability; Study population: humans and animal models; Publication period: January 1, 1989, to July 31, 2019; Language of publication: English and Spanish; Search terms: "Immunotherapy", "Neoplasms", "Mutation" and "Cancer Vaccines", and their equivalents in Spanish, which were combined with the "AND" and "OR" Boolean operators to establish the search equations.
Once the initial search was conducted, the following inclusion criteria were used to select studies for full analysis: studies addressing (i) the use of neoantigens predicted through a bioinformatic analysis; (ii) in vitro and in vivo immunogenicity assessment of protein derivatives, tumor lysate or RNA, and (iii) therapeutic effect of neoantigens in animal models of cancer or clinical response in humans to this type of therapy.
Results
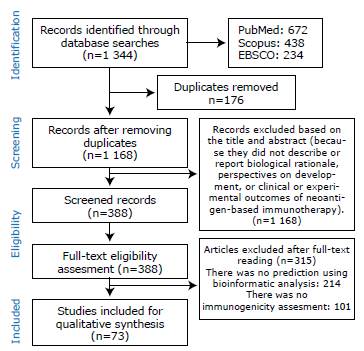
The initial search yielded 1 344 results (672 in PubMed, 438 in Scopus, and 234 in EBSCO), of which 176 were removed because they were duplicated and 780 were considered not relevant to the objective of the study or did not meet the inclusion criteria established after reading their titles and abstracts. A total of 388 articles were available for full-text review and eligibility criteria evaluation, of which 315 were excluded for not addressing the use of neoantigens predicted through bioinformatic analysis or not evaluating immunogenicity. 73 studies were finally included for full analysis (Figure 1).
Of the 73 articles included, 43 (58.90%) are original research works, of which 14 are clinical trials and 29 are experimental studies; of the latter, 10 were performed in in-silico models, 6 in vitro models, 2 in vivo models, and 11 used more than one experimental model. The remaining 30 studies were narrative reviews.
All articles were published in English, 47.94% were conducted in the USA, 43.83% in Europe, 5.47% in Asia and Oceania, and 7.73% in other American countries.
Table 1 presents the general characteristics of all the articles included in the review.
Table 1 General characteristics of the articles included in the review and relevance to its objective.
mRNA: messenger ribonucleic acid; DC: dendritic cells; RNA: ribonucleic acid; IDH1: isocitrate dehydrogenase type 1; IL-2: interleukin-2; HLA: human leukocyte antigen; NK: natural killer cells; MHC: major histocompatibility complex.
Source: Own elaboration.
Discussion
Personalized medicine increasingly aims to individualize disease prevention, diagnosis, and treatment according to the characteristics of each patient. In this context, progress in translational research (which are meant to benefit patients based on advances in basic research) and the results achieved in patients participating in clinical trials are critical inputs for planning new research and should serve as support for the development of treatments for cancer patients.
Advances in the study of tumor immunobiology have explained phenomena such as cancer immunoediting, a process that has allowed identifying neoantigens that escape tolerance induction and can be delivered to the immune system in the form of a vaccine so that they fight neoplasms.
Immunoediting in cancer
The theory of tumor immunoediting indicates that the immune system plays a decisive role in tumor control and development;27 it also describes cellular and molecular events that normally restrict tumor development and explains the appearance of cancer as a result of the escape of cells from immunosurveillance.
Immunoediting is defined by three key phases or events: elimination, equilibrium, and escape, which are described below:
Elimination
This is the phase during which the immune system (innate and adaptive) detects the transforming cells. This process is usually induced by carcinogens, radiation, viral infections, chronic inflammation and/or genetic mutations associated with the repression of intrinsic tumor suppression processes such as apoptosis, senescence, and tissue repair. These cells express danger signals and stress-induced ligands that, when recognized by the innate immune system, promote the release of pro-inflammatory cytokines that attract the cells of the adaptive immune system necessary to eliminate tumor precursors.28,29
Equilibrium
Some transformed cells survive the elimination phase and move on to the next phase, called equilibrium, where their proliferation is controlled. This phase, whose duration varies depending on the type of tumor, is not clinically evident; however, in it, the transformed cells determine their immunogenicity and influence the cellular context of the tumor infiltrate. If the latter is suppressive and the anergy of antitumor T cells is favored by the suppressive microenvironment of the tumor, then a state is created that favors the escape of transformed cells from the surveillance of the immune system, for example, the presence of myeloid-derived suppressor cells expressing CTLA-4 or the overexpression of molecules such as PD-L1 by the tumor cell, which are usually expressed as a mechanism of peripheral tolerance in non-tumor cells.
Escape
During the escape phase, the transformed cells evade recognition and elimination by the immune system and proliferate, and the tumor becomes clinically evident.31 Expression of apoptosis inhibitors, secretion of immunosuppressive cytokines such as TGF-β, and recruitment of myeloid-derived suppressor cells are mechanisms that facilitate this escape.
At the intrinsic level, tumor cells have escape mechanisms such as loss of expression of major histocompatibility complex class I (MHC-I) molecules.32 Although tumors are infiltrated by neoantigen-specific T cells,33 they do not exert their antitumor activity due to the suppressive microenvironment.
In 2017, Balachandran et al.33 demonstrated that long-term survival in patients with pancreatic cancer (a highly lethal tumor) depends mainly on T-cell infiltrate in tumors and on the quality (determined by a series of biophysical parameters such as homology determination with a microbial epitope) and not on the quantity of the neoantigen. This highlights the need to develop strategies that, on the one hand, favor the migration of neoantigen-specific T-cells to the tumor and, on the other hand, that the selection criteria of the latter take into account biophysical parameters that improve the likelihood of adequately stimulating T cells.
Therapies based on their immunological principle
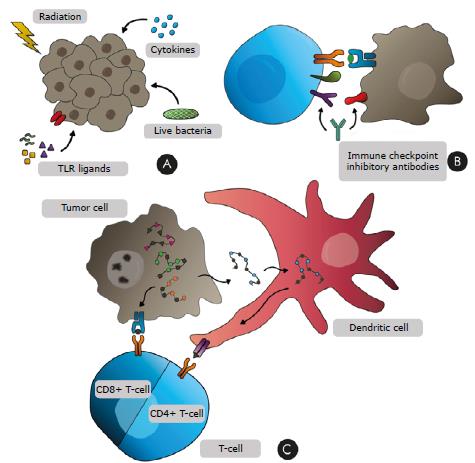
Tumor immunoediting theory has influenced the design of different immunotherapy modalities to counteract the suppressive effects of the tumor microenvironment, recover T-cell antitumor activity, and eradicate tumors already established according to three immunological principles, as shown in Figure 2:
Source: Own elaboration.
Figure 2 Therapies based on their immune principle. A) non-specific immune stimulation; B) immune checkpoint inhibitors; C) antigen presentation in the context of the HCM. LT CD4: CD4-positive T cell; LT CD8: CD8-positive T cell; TLRs: toll-like receptors.
Nonspecific immune stimulation with cytokines, toll-like receptor (TLR) ligands, radiation, and live bacteria have a pleiotropic effect, stimulate effector T cells, and inhibit regulatory T cells; some of these therapies have already been approved and used for the treatment of melanoma and renal carcinoma35-38 (Figure 2A). Monoclonal antibodies such as nivolumab and other immune checkpoint inhibitors,39 in turn, restore communication between antigen-presenting cells (APC) and T cells, and thus their cytotoxic potential40 (Figure 2B). Finally, personalized neoantigen-based therapeutic vaccines,41 the use of DCs as immunostimulants for the delivery of tumor-specific antigens (e.g., sipuleucel-T),42 and ACT seek to enhance antigen processing and presentation or to transfer in vitro-expanded tumor-infiltrating lymphocytes to the patient in order to restore tumor surveillance by these cells (Figure 2C).
From immunological theory to neoantigen-based vaccine design
Translational medicine requires the interdisciplinary cooperation of experts from several areas of biomedicine to successfully design therapeutic cancer vaccines.
The first step to establish personalized cancer therapies is to preserve the tumor so that the exome and transcriptome can be analyzed to identify mutations and possible neoantigens that are useful for designing vaccines43 (Figure 3); subsequently, the intratumor-al immune infiltrate should be evaluated to improve the prognostic value of the tumor. A tool that allows to evaluate how the immune system fights the tumor and helps choose a personalized treatment for colon and rectal cancer is called Immunoscore, which improves prognostic performance by 100 to 1 000 times compared to the TNM Classification of Malignant Tumours.
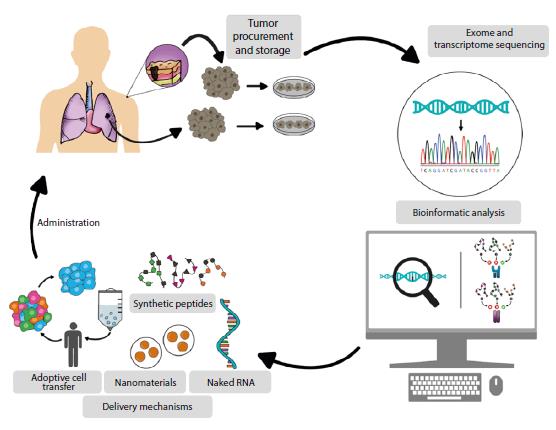
Source: Own elaboration.
Figure 3 Overview of the process of developing neoantigen-based vaccines for cancer immunotherapy.
A protocol consisting of the following steps should be used for the development of tumor-based neoantigen-based vaccines:
Tumor tissue collection and preservation
In general, therapy against the primary tumor is not effective against metastases due to tumor heterogeneity;32,33 therefore, primary and secondary tumor samples must be collected over different periods of time, as well as liquid biopsies, to obtain information, from a blood sample, on circulating tumor cells and tumor-derived cell-free nucleic acids, exosomes, and platelets.44
Bioinformatic analysis
Tumor exome and transcriptome sequences are established using state-of-the-art nucleic acid sequencing techniques and then compared with healthy tissue sequences (peripheral blood leukocytes obtained from a patient's blood sample) to determine the universe of tumor cell mutations. It is important to keep in mind that tumor RNA sequencing makes it possible to determine the levels of transcript expression that code for mutations.
After establishing the universe of mutations, these data are subjected to an in-silico analysis to obtain information on the interactions of the neoantigen with the patient's class I and class II human leukocyte antigen (HLA) molecules. This analysis also reveals parameters that allow the selection, among the universe of predicted neoantigens, of those most likely to stimulate the antitumor T cell response (both CD4 and CD8) in the patient.45
Identification and annotation of tumor-specific somatic mutations
Bioinformatic analysis is carried out in stages and the first is performed based on the raw data of the patient's normal and tumor exome sequences, which are text files in which sequenced DNA segments (reads) are recorded with their respective sequencing qualities and given a value for each sequenced nucleotide. The patient's normal and tumor exomes are reconstructed based on these data through a process called reference alignment46 in which each read is assigned a position with respect to a reference genome.
Once the exomes have been reconstructed, a comparison between normal and tumor exomes occurs during a second stage to identify somatic mutations.47 Finally, in a third stage, a mutation database search is performed to obtain information about each mutation (name of the gene where the mutation is located, transcripts associated with that gene, type of mutation, etc.)48 Additionally, the sequencing information provides a high-resolution HLA haplotyping used to determine HLA-restricted epitopes, as well as the stability of the neoantigen in each HLA haplotype molecule expressed by the patient.
Tumor tissue transcriptome analysis
The sequencing of a transcriptome is similar to that of an exome, both in the way they are processed and in the type of data obtained. This type of analysis also starts from text files containing the sequenced reads with the respective sequencing quality values and is done by aligning the sequencing reads with a reference genome.46,49 Then, using databases containing the names and sequences of known transcripts, the transcripts encoded in the patient's transcriptome are identified, and its expression levels identified are quantified through the number of reads associated with each transcript. This quantification is usually presented in units known as FPKM (fragments per kilobase of transcript per million mapped fragments).50
Epitope prediction
The candidate epitopes capable of being presented on the patient's MHC molecules should be established based on the tumor cell-specific mutations identified in the genome and the expression level determined by the transcriptome. This prediction is based on in-silico tools that estimate the splicing of the peptide by the proteasome, as well as its presentation according to its affinity and stability in the peptide-MHC complex.51-53 These tools are based on artificial intelligence methods, such as neural networks, that evaluate possible paired combinations between the set of epitopes that could be generated around each mutation.
Finally, in conformity with selection criteria such as experimental binding affinity values (IC50), FPKM, the rate of change between the IC50 of both the mutated peptide-MHC complex and the wild-type peptide- MHC complex, the comparison of the half-life of both peptide- MHC complexes (native and mutated), and a small number of candidate epitopes, whose immunogenicity should be evaluated in vitro, is prioritized.45
Alternative formulations of neoantigens to ensure delivery to the immune system
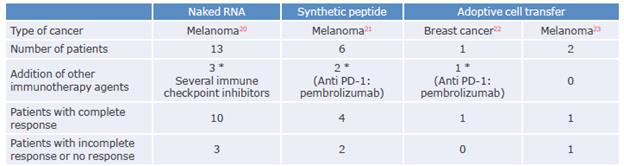
Once the neoantigen has been selected, a vehicle that makes an efficient and safe delivery of the neoantigen to the immune system should be chosen.54 To this end, several strategies have demonstrated significant clinical results (Table 2).
Naked RNA
Research on the use of naked messenger RNA (mRNA) as a source of antigen has been aimed at achieving the stability of the RNA necessary for the correct uptake and translation by APCs. In this process, genetic engineering has allowed to incorporate changes in RNA that increase its bioavailability for DCs, which are the cells that process and present antigens more efficiently. The uptake mechanism of injected mRNA is not entirely clear; however, it is known that it is rapid, not very saturable and dose and temperature dependent, and some authors suggest that macropinocytosis may be the mechanism employed.55,56
Changes made to mRNA to achieve the desired stability include the modification of the 3'UTR and 5'UTR regions structure, the use of mRNA 5' cap analogs, and the addition of nucleotide flanking sequences that favor their mobilization through endoplasmic reticulum membranes and antigen presentation mediated by MHC molecules.25 Also, since the mRNA delivery site affects its uptake by DCs, mRNA should, ideally, be injected directly into the lymph nodes or the soft tissue near the nodes.57
The prophylactic and therapeutic response of mRNA vaccines has been demonstrated in both animal models57 and in phase I and II clinical trials with human patients with melanoma20,58,59 and renal cell carcinoma.60 Many of these assays use tumor-associated antigens that have validated the viability and safety of this therapy.59,60 For example, Sahin et al.,20 in a study based on the use of mRNA vaccines coding for tumor mutanome neoantigens, reported that an antitumor response was obtained in 10 of 13 patients with metastatic melanoma.
Synthetic peptides
The rise of peptide neoantigen vaccines is related to the high degree of understanding of how peptides processed and presented in MHC molecules on the APC surface mediate the T cell activation involved in immunity against infectious agents and cancer.61
Some of the advantages of using peptides as antigens are the relative ease of synthesis, low production costs, low toxicity, and proven stability under temperature changes.54
It should be noted that, in order to generate long-term protective responses, this type of vaccine has been administered together with adjuvants, which are usually TLR agonists such as Poly-ICLC (TLR3 agonist) and have the function of improving immunogenicity and promoting the delivery of the neoantigens to the immune system.62,63
Given this scenario, representative studies have been conducted in which patients with different types of cancer are vaccinated with short and long synthetic peptides (9 to 27 amino acids, respectively) containing a tumor-specific mutation, for instance, a variation of the enzyme IDH1,64 or others obtained from sequencing widely studied tumor cell lines such as B16F10 melanoma,26,65 MCA sarcoma,64 or MC38 colon carcinoma,66 which are used in different mouse models of cancer where strong in vitro immunogenicity and in vivo antitumor response have been demonstrated, mostly by CD4 + 26,64,65 and CD8+ T cells.64,66
In humans, Ott et al.21 conducted a study that showed that 4 of 6 melanoma patients tested (4 stage IIIC without metastases and 2 stage IVM1/b with metastases) achieved complete remission following neoantigen vaccine-only treatment. This study generated peptides of 15 to 30 amino acids that were administered by subcutaneous injections into the limbs, creating an antitumor response mediated by polyfunctional CD4+ and CD8+ T cells21 (Table 1). Similarly, several phase I clinical trials are currently under development67,68 to replicate the success of this treatment in melanomas.
Synthetic peptides derived from neoantigens can be used to stimulate in vitro DCs obtained from peripheral blood mononuclear cells and then delivered to the patient as adjuvant autologous therapy. These DCs process and present the selected peptide sequence in the MHC and acquire the ability to efficiently activate cellular-mediated immune responses carried out by CD8+ T cells against the tumor.69
Adoptive cell transfer
This type of therapy consists in the culture and in vitro or ex vivo amplification of T cells extracted from the patient's tumor or peripheral blood (autologous) or from T cells extracted from a donor (allogeneic) individual. These samples are then transferred to the patient to exert cytotoxic activity against the tumor.70
The T cells to be transferred are usually selected from the total repertoire of cells extracted according to the reactivity when cultured with tumor antigens from cell fragments, cell lines, suspensions, lysates, or synthetic tumor proteins.71
The reactivity of the T cells to be transferred is often demonstrated by quantifying the level of cytokines using enzyme-linked immunosorbent assays, while flow cytometry is used for classification and selection, thus characterizing cells according to relevant cell biomarkers such as 4-1BB, expressed by activated CD4+, CD8+ and CD45+ cells in response to neoantigen stimulation.72,73
In this therapy, T cells that recognize mainly the neoantigen without any activity against the sequence of the wild-type peptide are preferred to avoid autoimmunity.74,75 This strategy has demonstrated significant clinical responses, ranging from increased overall survival to complete regression of metastatic tumors in breast cancer, melanoma, among others22,76-78 (Table 1).
Future challenges and prospects
Advances in various immunotherapy modalities raise relevant questions whose answers can be helpful to understand the singularities of the immune response and the mechanism of action of these therapies in each type of tumor explored. In this regard, research in the coming years should focus on replicating the results of previous studies and overcoming the most significant challenges that have arisen in different populations and cancer histotypes. Some of these challenges are presented below.
Adjuvants and antigen delivery
In order to increase immunogenicity and promote an optimal response, immunotherapy is combined with agents that simulate the first steps in the recognition of an infection, mainly TLR agonists.36 However, there are other molecules that seek to improve antigen delivery and are under study; these include monoclonal antibodies that target tumor antigens to DCs and activate them, such as anti-CD20579 in CDX-1401, which significantly increases the effectiveness of antigen presentation.
Differential mutational load
Most studies that have analyzed the immunogenic potential of mutations in cancer treatment have been done on the basis of identification of single-nucleotide variants (SNV); this has made it possible to establish that melanoma is among the cancers with the highest proportion of SNV, so the first clinical trials were performed on this type of tumor. However, it is important to keep in mind that other types of mutations also have immunogenic potential; for example, insertion/deletion mutations, which modify the reading frame to a greater extent have been shown to generate more neoantigens per mutation and have a higher capacity to bind to MHC-I.80,81
This is supported by some proteogenomic analyses that have demonstrated the importance of considering the generation of neoantigens from non-conventional alternative reading frames (cryptic peptides), since it was proved that they constitute 10% of all peptides generated that can be presented in the context of MHC-I, but that they have been omitted in many studies due to of their non-canonical nature.82
Immune evasion
The processes that lead to generating resistance to immunotherapy are related to mechanisms intrinsic to each therapeutic strategy. For example, in tumor immunoediting, somatic mutations may occur in neoantigens and possibly in the MHC,23,83 which may explain inefficient responses and treatment failures in some clinical trials84 since the functional state of these molecules is being modified (both neoantigens and MHC), destabilizing the interaction of the peptide-MHC complex.
HLA mutations have been characterized as "soft" or "hard" depending on the feasibility of reversing them85 lesions derived from hard mutations generate more serious and irreversible immune response escape routes associated with metastatic progression and/or tumor recurrence, while those derived from soft mutations are more easily correctable and often associated with epi-genetic and reversible changes with treatment.
Likewise, strategies for dealing with HLA alterations are different in each case;86 for example, epigenetic regulation using the histone deacetylase inhibitor AR-42 is useful in mice to restore MHC-I expression,87 whereas recombinant adenoviruses loaded with missing genes have been used for hard mutations, as in the case of mutations in β-2 microglobulin88 or the transfer of natural killer cells preactivated with a specific cocktail of cytokines,89 obtaining good results.
Detection of HLA mutations is complex for several reasons. On the one hand, these gene sequences are highly polymorphic, and on the other, the reference genomes most used to determine them are limited. Given the situation, a method was recently developed to analyze this type of mutation more effectively, which also allows to identify its recurrence in different types of cancer.
Redefinition of the concept of neoantigen
There is another type of antigens recognized by T cells that escape deletion in the thymus and are encoded in pro-to-oncogenes responsible for inducing post-translational modifications in proteins that promote metabolic alterations and changes in tumor transcriptome. These proteins, which are abnormally glycosylated or phosphorylated, have not been studied at the same depth as a source of neoantigens for cancer treatment; nevertheless, they are highly immunogenic and responsible for a potent antitumor response, as demonstrated by Brentville etal.24 in a recent study, in which they found that antigens generating high reactivity are citrullinated self-peptides.24
It is striking that, in several clinical trials, the predominant response against neoantigens was dependent on CD4+ T cells,20,21,78 in which in some cases it was possible to demonstrate their cytotoxic capacity.24 This undoubtedly calls for much more attention to the helper-like response of T cells, which is mediated by MHC-II molecules in the selection and screening of neoantigens for clinical application.91
Consensus, efforts, and paradigms
Genomic heterogeneity between and within tumors and their ability to escape immune pressure make cancer treatment a major challenge; therefore, a consensus on their theoretical and therapeutic approach is desirable. In this sense, the interdisciplinary cooperation of biomedical professionals to obtain the tumor and to carry out the corresponding bioinformatic analysis, together with the combination of therapies with different immunological principles, play an important role that should not be ignored.
For example, in Colombia, the Immunology and Trans-lational Medicine group of the Universidad Nacional de Colombia has worked for more than 14 years on the advancement and implementation of clinical and preclinical studies in immunotherapy against cancer and is currently conducting the first clinical study in the country on immunotherapy based on neoantigens in patients with triple-negative breast cancer using autologous DC as adjuvant therapy.
These studies, in which different bioinformatic platforms have been articulated, have allowed to establish the universe of mutations present in patients with breast cancer and to identify tumor cell-specific neoantigens with the potential to be used as neoantigens in the clinical phase of the study. However, the process of in vitro evaluation of the response of specific T cells against these neoantigens has been a limitation given the low frequency of the repertoire of these cells against predicted antigens. Even so, it is expected that with the use of autologous peptide-pulsed dendritic cells, the specific T-cell clones against the neoantigens used will increase.
Nevertheless, personalized medicine worldwide demands a change in the paradigm of organizing, regulating, and following up clinical studies (originally designed for drug development) since there is a clear need to streamline the evaluation of clinical studies in which personalized vaccines based on neoantigens are investigated and in which the investigational agent is not a drug.92
Conclusions
Personalized immunotherapy with tumor-based neoantigen vaccines is becoming an excellent alternative for treating cancer worldwide. However, its proper and massive implementation faces some technical and theoretical challenges that must be overcome, such as the complexity of the analysis of neoantigens and its in vitro validation, inadequate conservation of tumors for study, and the high cost of obtaining peptides with clinical quality and effective adjuvants for human use. It is thus necessary to use it in combination with conventional treatments, generate more knowledge that will help clarify the immunobiology of cancer, and reduce the costs associated with its production.