Introduction
An acute lower respiratory tract infection (ALRTI) affects the lower respiratory tract and involves the trachea, bronchi, bronchioles and alveoli, resulting in tracheitis, bronchitis, bronchiolitis, and pneumonia, respectively.1 Its spectrum of clinical presentation is wide and ranges from asymptomatic cases to patients with severe restrictive and obstructive respiratory diseases requiring multiple empirical therapies such as antibiotics, even for viral infections.2 ALRTIs are common and potentially life-threatening in children, occurring most frequently in early childhood, an age when the immune and respiratory systems are still immature.3
ALRTIs are among the three leading causes of death and disability in the pediatric population. Worldwide, it is estimated that almost 4 million deaths per year are associated with this infection, and it is also the leading cause of death in children under 5 years of age.4 In Latin America, between 140 000 and 150 000 children under 5 years of age die each year from an ALRTI, of whom 100 000 are under 1 year of age and between 40 000 and 50 000 are between 1 and 4 years of age.5 In Colombia, ALRTIs are the fifth cause of morbidity and mortality in the general population; they have a seasonal pattern that spans two periods, one between March and June and the other between September and December. In 2019, the country's mortality rate for this cause was 11.14 cases per 100 000 children under the age of 5.6
The etiology of ALRTIs in children under 5 years of age is predominantly viral,7 with respiratory syncytial virus (RSV), adenovirus (ADV) and influenza type AH3 being the most frequent causes.8 On the other hand, the bacteria most frequently detected in ALRTI cases are Streptococcus pneumoniae, Haemophilus influenzae type b, Bordetella pertussis, Chlamydia trachomatis, and Mycoplasma pneumoniae.8
In Colombia, the implementation of immunization or vaccination in children under 5 years of age, which includes protection against S. pneumoniae, H. influenzae, and B. pertussis,9 has made it possible to reduce mortality from respiratory infection in this population.10
It has been established that viruses detected in the nasopharynx are almost always a cause of respiratory infection, while bacterial growth in the trachea may be due to specimen contamination with organisms colonizing the upper respiratory tract.11 Colonization is understood as a sufficient concentration of organisms that can be detected but do not cause any signs and symptoms, whereas infection corresponds to the detection of microorganisms accompanied by signs and symptoms resulting from the inflammatory process or contamination. Consequently, the factor that determines whether a patient is colonized or infected is their signs and symptoms.11
In general, rapid screening techniques, cultures of respiratory specimens, and blood cultures are commonly used for ALRTI diagnosis, although these tests can be inaccurate and untimely because they detect a limited number of microorganisms.7 However, the development of molecular techniques has enabled the detection of multiple pathogens using a single molecular assay with high sensitivity, specificity, and ease of access,12,13 thus transforming the paradigm of the single causative agent by demonstrating that there are multiple combinations of causative agents that can modify the natural course of an ALRTI.14 One example is the detection of mixed infection associated with prolonged hospitalization and poor clinical prognosis.15 It should be noted that in the present study, coinfection was defined as an infection by two or more viruses and mixed infection as an infection caused by a variety of microorganisms (virus-bacteria).16
The gold standard in the detection of respiratory viruses is the real-time multiplex polymerase chain reaction (RT-PCR) technique, in which samples of infected cells taken from a nasopharyngeal swab are analyzed.17 This assay, which was used to develop the present study, makes it possible to identify the genetic material of the viruses and bacteria found in the nasopharynx and, therefore, its use in primary care allows for the prevention of the transmission of these microorganisms and the reduction of bacterial resistance through the rational use of antibiotics.18,19 Furthermore, the detection of infections caused by single agents, coinfections, and mixed infections with this technique16 could predict the modulation or the exacerbation of the inflammatory response; an example of this paradigm shift (ARI caused by a single agent) is the phenomenon of viral interference, in which a virus competitively suppresses the replication of other infecting viruses and modulates virulence and cell death by altering cellular immune response and the severity of the ALRTI.16
The purpose of this study was to characterize the pathogens identified by RT-PCR in nasopharyngeal swabs, as well as clinical variables and laboratory findings in children under the age of 5 who were diagnosed with ALRTI and hospitalized in Bogotá, Colombia.
Materials and methods
Type of study and study population
Cross-sectional study in which microorganisms were identified using the RT-PCR technique in nasopharyngeal swab samples from children under 5 years of age diagnosed with ALRTI, hospitalized between September 2019 and March 2020 at the Clínica Cafam in Bogotá, Colombia, and who met the following inclusion criteria:
Signs of systemic inflammation: heart rate >180 bpm, tachypnea (respiratory rate >60 rpm in children under 2 years old and >50 rpm in children over 2 years old), and temperature >38°C.
Clinical signs of lower respiratory involvement (wheezing, rhonchi, or rales) or shortness of breath (retractions or nasal flaring).
Peripheral oxygen saturation measured by pulse oximetry <90% indicating the need for oxygen therapy.
Diagnosis of bronchiolitis, recurrent wheezing or pneumonia at the discretion of the attending emergency physician.
In-hospital treatment requirement.
Informed consent form signed by parents or legal guardians.
Patients with a history of chronic diseases, immunodeficiencies, prematurity, hospitalization in the two months prior to the study period, and positive asthma predictive index were excluded because respiratory exacerbations of non-infectious etiology can be found in these cases. Thus, the sample was consecutive and non-probabilistic, and consisted of 81 patients.
All the children were admitted by the emergency department and were evaluated by the on-call attending physician following a thorough physical examination that included a review of vaccination status, blood count, measurement of C-reactive protein level, and chest X-ray. Based on this evaluation, the physician made the diagnosis of ALRTI and ordered hospitalization in the clinic as well as the start of treatment at their discretion.
Procedures
All 81 children underwent nasopharyngeal swabbing with nylon swabs (COPAN, Italy) that were stored in universal transport medium tubes (UTM MR, COPAN, Italy).
Samples were taken to a reference laboratory in the city (COLCAN), where the multiplex RT-PCR Allplex® assay (Seegene, South Korea) with one-step reverse transcription was performed to detect, using AllplexTM 1, influenza virus type A, influenza virus type B, RSV type A (RSVA) and RSV type B (RSVB). AllplexTM 2 was used to detect ADV, human metap-neumovirus (HMPV), enterovirus, and parainfluenza virus (1, 2, 3, and 4). AllplexTM 3 was utilized to detect human bocavirus (1, 2, 3, and 4), rhinovirus (RVA, RVB, and RVC), and coronavirus (HCoV-NL63, HCoV-229E, and HCoV-OC43). Finally, AllplexTM 4 was used to detect H. influenzae, M. pneumoniae, Chlamydia pneumoniae, Legionella pneumophila, S. pneumoniae, B. pertussis, and Bordetella parapertussis.
The Allplex® multiplex RT-PCR assay uses MuDT (multiple detection temperatures) analysis technology, which allows detecting multiple targets in a single fluorescence channel without melting curve analysis, and the TOCETM (Tagging Oligonucleotide Cleavage and Extension) technique, which utilizes indirect signal generation by using two components (pitcher and catcher) to design oligonucleotides to detect the DNA target. During each cycle, the pitcher, a dual-purpose oligonucleotide that binds specifically to target genetic material, is cleaved to release an unlabeled extender molecule that serves as a primer for an artificial template, which is a quenched-fluorescent molecule, also known as the catcher. The DNA and the extension of the extender in the receptor generate a fluorescence signal that directly correlates to the amount of target DNA.20
RT-PCR assay results were classified as mixed infection when it was a virus-bacteria result or as coinfection when it was a virus-virus result.16 When no viruses were detected it was classified as none and when a single microorganism was found, it was defined as single pathogen.
In the present study, bronchiolitis was defined as the first episode of wheezing in patients younger than 2 years; recurrent wheezing, as episodes of wheezing following bronchiolitis; and pneumonia, as signs of lower respiratory tract involvement associated with pulmonary consolidation on chest X-ray.
Data on clinical variables, radiological findings and laboratory results were obtained by reviewing the participants' medical records.
Statistical analysis
In the descriptive analysis, absolute and relative frequencies were used for categorical variables, while measures of central tendency and dispersion were used for quantitative variables according to the distribution of the data, as determined by the Shapiro-Wilks test.
Demographic variables (age, sex, and age group) and clinical variables (physical examination, laboratory results, radiological findings, microorganisms identified, oxygen therapy, antibiotic treatment, referral, and intensive care management) were analyzed. In addition, a graphical descriptive bivariate analysis between the RT-PCR assay results and the following variables was performed: age group (young infant: 3-12 months, old infant: 13-24 months, and preschooler: 25-60 months), clinical characteristics (physical examination findings detected by the emergency physician and described in the medical record), laboratory results (blood count and C-reactive protein levels), and radiology reports (pneumonia, atelectasis, normal, etc.)
Finally, Spearman's correlation coefficient was used to determine the correlation between the number of microorganisms per patient, on the one hand, and the blood cell count and C-reactive protein levels, on the other. All statistical analyses were performed in R v4.0 TM statistical software and a significance level of p< 0.05 was considered.
Ethical considerations
The study took into account the ethical principles for research involving human subjects established by the Declaration of Helsinki21 and the provisions on health research of Resolution 8430 of 1993 of the Colombian Ministry of Health.22 The ethical guidelines for biomedical research of the Council for International Organizations of Medical Sciences were also followed.23
As mentioned in the inclusion criteria, informed consent was required from all patients' parents. The research project was evaluated and endorsed by the Institutional Research Ethics Committee of the Clínica Cafam according to Minutes No. CIEI CAFAM/2020-5884 of August 12, 2020.
Results
The mean age of the participants was 17.23 months (±14.44) and 54.32% were male. The most frequent age group was young infant (51.85%). The most frequent ED diagnosis was recurrent wheezing and bronchiolitis, with 37.04% each.
A total of 149 microorganisms (60.40% viruses) were identified in 63 children (77.78%); mixed infection was identified in 48.15% of the cases and no microorganism in 22.22%. The most frequent viruses were RVA (14.48%) and RSVA (11.03%). The most frequent bacteria were H. influenzae (20%) and S. pneumoniae (17.93%).
During hospitalization, there were 83 episodes that required oxygen support by different routes; 78 of these requirements were for nasal cannula, 3 for Venturi mask, and 2 for high-flow cannula. Regarding treatment, it was found that 17 children were treated with antibiotics, being penicillin G sodium the most frequently used (82.30%); moreover, 41.10% of the 14 patients who had a viral result in the RT-PCR received antibiotics. Of all participants, 6.10% received oseltamivir, 30.8% were referred to higher level of care, and 3.77% required pediatric intensive care unit management (2 of these patients had coinfection). The clinical and demographic characteristics of the study population are presented in Table 1.
Table 1 Demographic and clinical characteristics of patients diagnosed with acute lower respiratory infection.
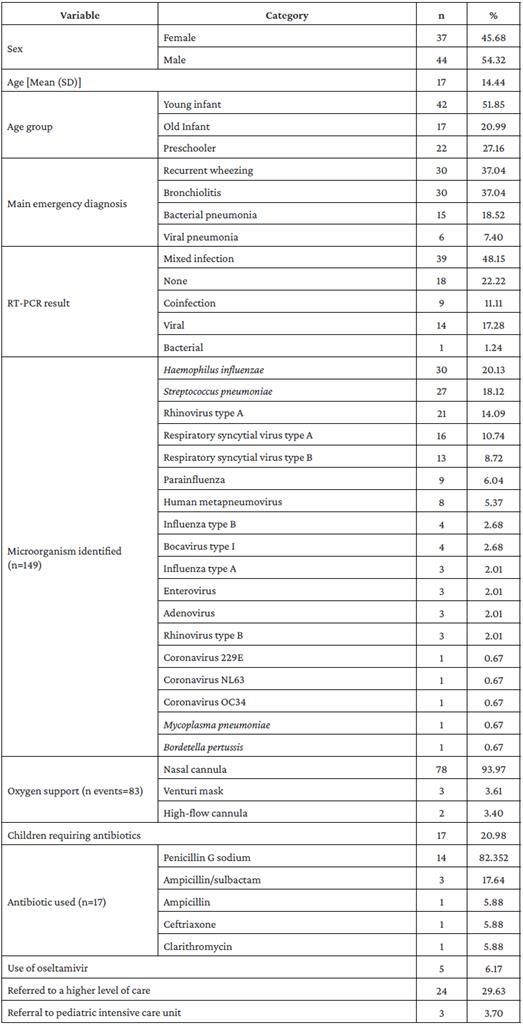
SD: Standard deviation.
Source: Own elaboration.
Table 2 shows that at least one microorganism was identified in 63 patients (77.78%). In addition, according to diagnosis, the following results were observed:
In patients with bronchiolitis, no microorganisms were found in 13.33% (n=30); the most frequent combination was RSVA, H. influenzae and S. pneumoniae, with 10%, and the most frequent pathogen was RSVA, also in 10% of the cases.
In patients with recurrent wheezing (n=30), no microorganism was found in 26.77%. The most frequent combinations of microorganisms, with 6.67% each, were HBoV1, RVA, H. influenzae and S. pneumoniae; RSVA and S. pneumoniae; RVA and S. pneumoniae; and RVA and H. influenzae. The most frequent single pathogen was HMPV, also with 6.67%.
In patients with viral pneumonia (n=6), no microorganisms were found in 33.33%, and the most frequent combination of microorganisms was RSVB, RVA, and H. influenzae, with 6.67% each.
Finally, in patients with bacterial pneumonia (n=15), no microorganisms were found in 13.33%, while 66.67% had multiple pathogens, and the most frequent single pathogen was RSVB, with 20%.
Table 2 Frequency distribution of microorganisms identified by real-time multiplex polymerase chain reaction assay according to ED diagnosis.
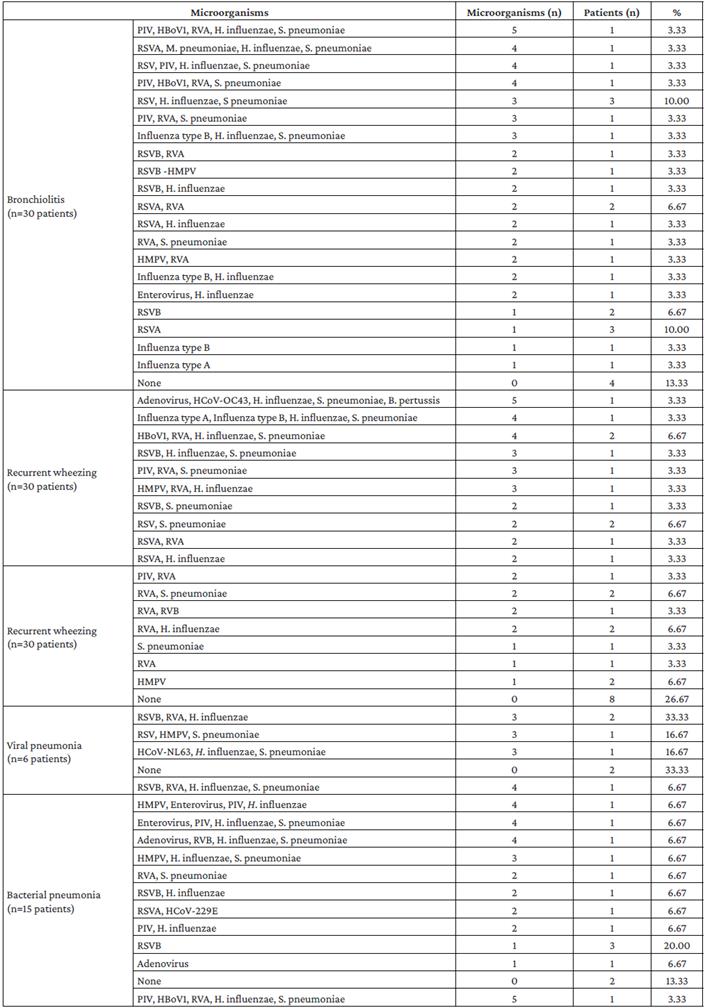
PIV: parainfluenza virus; HBoVl: human bocavirus type 1; RVA: rhinovirus type A; RSVA: respiratory syncytial virus type A; RSVB: respiratory syncytial virus type B; RVB: rhinovirus type B; HMPV: human metapneumovirus; HumanHCoV-OC434: human coronavirus OC434; HCoV-NL63: human coronavirus NL63; HCoV-229E: human coronavirus 229E.
Source: Own elaboration
Figure 1 shows that 75 microorganisms were identified in young infants, 64.00% of which were viruses, the most frequent being RVA (17.30%) and RSVA (16.00%); likewise, the most frequently identified bacteria in this age group were S. pneumoniae (18.70%) and H. influenzae (17.30%). On the other hand, 43 microorganisms were identified in old infants, 53.40% of which were viruses, the most frequent being RVA, RSVA, RSVB, and ADV (7% each). Finally, 31 microorganisms were detected in preschoolers, 61.20% of which were viruses, the most frequent being RVA (16.10%), parainfluenza (9.70%), RSVB (6.50%), RVB (6.50%), and HMPV (6.5%); the bacteria identified in this age group were H. influenzae (22.60%), S. pneumoniae (12.90%), and M. pneumoniae (3.20%).
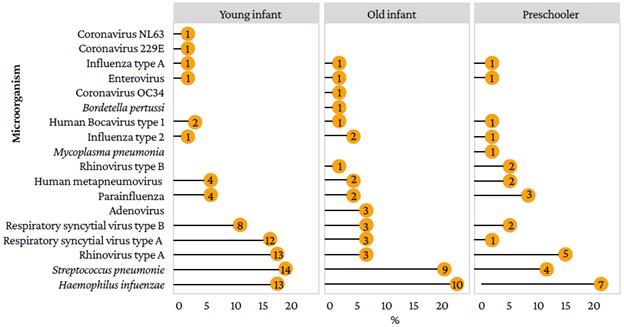
Source: Own elaboration.
Figure 1 Viruses and bacteria detected by real-time multiplex polymerase chain reaction assay according to age group.
Figure 2 shows that shortness of breath was higher in patients with mixed infection (32.98%) and with no microorganism (32.00%), that upper respiratory tract obstruction was important in cases of no microorganism (16.00%), that cough was higher in participants with coinfection (64.41%) and single pathogen infection (27.78%), that pharyngitis (22.00%) and conjunctivitis (8.00%) were more frequent in patients with no microorganism, and that fever was frequent in cases of mixed (29.79%) and single pathogen (18.52%) infection.
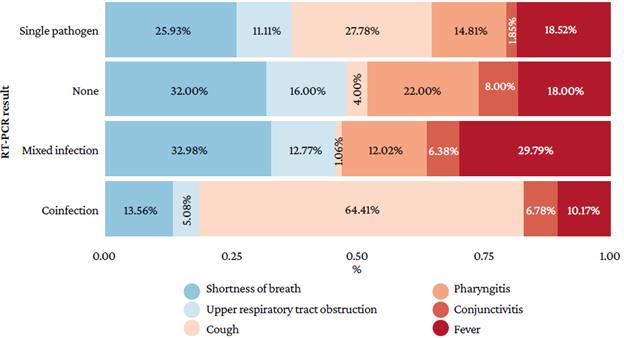
Source: Own elaboration.
Figure 2 Clinical findings in participants according to the real-time multiplex polymerase chain reaction assay result.
Figure 3 indicates that the lack of radiological findings was more frequent in cases of mixed infection (50.70%), that interstitial opacities were more frequent in cases of coin-fection (44.44%), and that consolidation was predominant when no microorganisms were found (23.33%). In addition, atelectasis was observed in cases of mixed infections (2.82%).
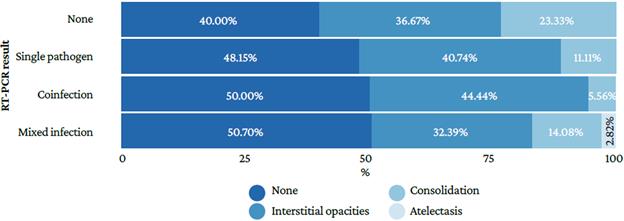
Source: Own elaboration
Figure 3 Radiological findings in the participants according to the multiplex real-time polymerase chain reaction assay result.
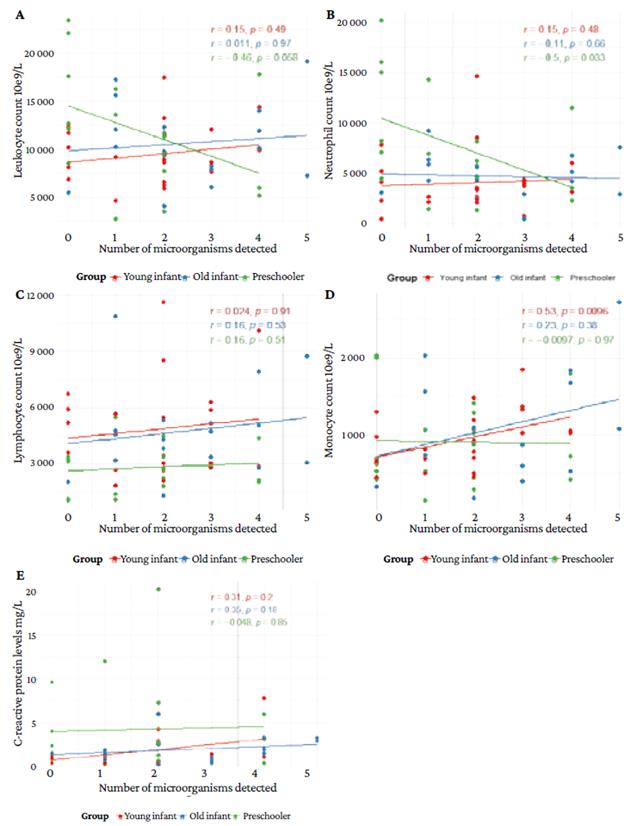
Figure 4 shows that blood cell and C-reactive protein levels are similar in all types of RT-PCR assay results.
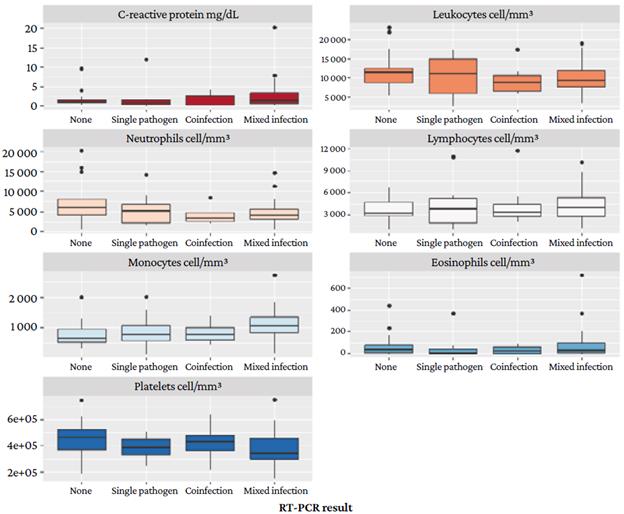
Source: Own elaboration.
Figure 4 C-reactive protein and blood cell levels according to the multiplex real-time polymerase chain reaction assay result.
Respiratory pathogens in children
Figure 5 shows a correlation between blood cell count and the number of microorganisms detected. Figures 5A and 5B show a negative correlation between the number of leukocytes and neutrophils and the number of microorganisms detected in patients in the preschool group, the latter correlation being statistically significant (r=-0.46; p=0.058 and r=-0.51; p=0.033). Figure 5C shows no significant correlation between the number of lymphocytes and the number of microorganisms (p> 0.05). Figure 4D shows a positive correlation between monocytes count and the number of microorganisms in young infants, being this correlation statistically significant (r=+0.53; p=0.0096). Figure 5E shows no correlation between the level of C-reactive protein and the number of microorganisms in any of the groups (p>0.05).
Discussion
Generally, infectious diseases are attributed to a single pathogen, whose syndromic correlation provides an estimated diagnosis probability and, as a result, allows for decisions regarding initial empirical antimicrobial therapy.16 However, it has been reported that multiple infectious agents could be involved, but the clinical, diagnostic, and therapeutic implications of this situation are still unknown.24 This has been made evident by the development of multi-detection molecular techniques, which are characterized by improved operational performance.14 The RT-PCR assay, for example, searches for viral and bacterial genomes in a single reaction and reports the presence of multiple causative agents in less than 2 hours.16
In the present study, the majority of participants were male (54.32%) and infants (old and young) (72.84%), which coincides with what has been described in the literature.25,26 The latter finding is highly relevant since infants are at high risk of developing lower respiratory infections due to factors such as relative immature immune response and incomplete vaccination schedules, and also because this population usually attends daycare centers and kindergartens, places where various viruses circulate.27
The usual clinical presentation of ARLTI in the study population was obstructive, with bronchiolitis and recurrent wheezing being the most frequent diagnoses (37.04% each), manifestations usually attributed to viral infections.28 For its part, the prevalence of infection with two or more pathogens reached 59.26% (mixed infection or coinfection), a figure that is in the range described in the relevant literature, which ranges from 7% to 76% in children under 5 years of age.14,29-31
Diagnostic suspicion of ARLTI is usually confirmed on the basis of semiology and imaging findings. However, in the present study, there was a high heterogeneity among these clinical findings, preventing the identification of respiratory syndromes traditionally described in this population group, such as RSV bronchiolitis or bacterial pneumonia due to S. pneumoniae. This is in agreement with what Krause et al.19 reported in a literature review, in which no differences between clinical findings and pathogens identified in children with respiratory tract infections were observed.
Similarly, in the present study, there were also no significant differences in blood counts and C-reactive protein levels according to the RT-PCR assay result (no pathogen, single pathogen, coinfection, mixed infection), a finding similar to that reported by Lin et al.14 in a study of 474 samples from Taiwanese children with respiratory symptoms and clinical suspicion of virus infection, in which they found no significant differences according to the RT-PCR assay result (co-infection, single pathogen, and no infection) (p=0.227).
Moreover, viral infections were clinically unpredictable, and patients presented with fever, shortness of breath, and upper or lower respiratory tract obstruction, symptoms typically reported in the literature14,32,33 and which may be mistaken for a bacterial infection.
On the other hand, in the present study, 41.10% of the patients who had a viral result in the RT-PCR assay received antibiotic therapy, so the relevance of the result of these tests to support clinical decision should be cautious, considering that there may be microorganisms in the upper respiratory tract that do not necessarily explain the lower tract infection (i.e., ARLTI). In addition, this decision should be consistent with all the evidence provided by the clinical manifestations of the patient and the laboratory results to avoid unnecessary use of antibiotics and the induction of bacterial resistance.34
As mentioned above, when analyzing the RT-PCR assay results, it was found that there were cases of mixed infection and coinfection. In this regard, it has been established that although bacteria that normally inhabit the mucosa of the upper respiratory tract can be detected in mixed infections, virus-bacteria interaction has been associated with successful invasion and colonization of pathogens in the human body.11 However, Meskill et al.,35 in a recent literature review, reported that even though the evaluation of the impact of viral-bacterial coinfection on disease severity is an advancing field of study, there are no studies that report a decrease in severity with this type of coinfection. In this sense, the present study found that 2 of the 3 patients who required management in the pediatric intensive care unit had a mixed infection.
In the present study, the most frequent combination of microorganisms in cases of bronchiolitis was RSVA, H. influenzae, and S. pneumoniae (10%). In this regard, it has been described that the detection of RSV and S. pneumoniae is significantly associated with severe disease35 and that it has been linked to seasonal increases in pneumococcal disease.36
Of the 149 microorganisms identified, the most frequent virus was RVA (14.09%), followed by RSV (Type A: 10.74% and Type B: 8.72%). Rhinovirus is a rapidly replicating virus that can interfere with the replication of other viruses and is the second most common causative agent of bronchiolitis, after RSV, in hospitalized patients;35 nevertheless, since many patients with this virus are asymptomatic, molecular diagnostic tests are not usually performed to detect it due to its high prevalence in asymptomatic patients.35 For its part, RSV is characterized by high viral loads and is the main cause of hospitalizations in children between 1 and 3 years of age.37
The literature describes that the most frequent type of interaction in cases of coinfection is viral interference, in which the virus competitively suppresses the replication of other coinfecting viruses.16,35 Similarly, in other cases, as reported by Kumar et al,16 coinfections have no effect on virus replication and, therefore, all coinfecting viruses can coexist (accommodation). Likewise, it has been reported that coinfections can modulate or exacerbate viral virulence38 and cell death, thus altering the severity30 and epidemiology of the disease.16
The host is a fundamental part of this interaction (coinfection), which was evidenced in the present study by the correlation between the number of microorganisms and the number of activated or inactivated cells during the inflammatory process according to age group. The negative correlation in the preschool group between the absolute number of leukocytes and neutrophils and the number of pathogens detected (r=-0.46; p=0.058 and r=-0.51; p=0.033), as well as the positive correlation between the monocyte count and the number of microorganisms identified in young infants (r=+0.53; p=0.0096), is striking. Although no studies in children that evaluated such correlations were found, these results could be explained by the fact that respiratory viruses may impair neutrophil function, decreasing oxidative burst and enhancing neutrophil apoptosis, which increases the susceptibility to bacterial superinfection.35 In turn, the interaction between pathogens (virus-bacteria) could alter monocyte function, thereby decreasing cytokine production and activity, as well as the proper functioning of immune response.39
Considering the results of the present study, it is possible to state that there is still much to learn about respiratory infections in the pediatric population caused by multiple microorganisms and the usefulness of their detection in clinical practice. Thus, it is clear that, at present, the concept of a single causative agent of ALRTI cannot be considered in a generalized manner in the care of these patients since the detection of multiple pathogens by new diagnostic techniques, which are becoming increasingly common and affordable in the hospital setting, may require the adjustment of therapeutic interventions in this population.
The limitations of the present study include the fact that it was not possible to follow up the patients in the medium term or to determine the type of infection or colonization of the nasopharynx by means of a gold standard test, and that there was not a sufficient sample to perform statistical inference and association with serious outcomes. Furthermore, due to the fact that not all the agents identified are pathogenic, as they may be colonizers or co-infectants, it is recommended that further research quantifies viral loads or additional cultures to establish a pathological correlation.
Conclusion
Multiplex RT-PCR is a diagnostic test that allows the detection of genetic material of microorganisms that are infecting or colonizing an individual. In the present study, it allowed identifying microorganisms in most children, as well as cases of mixed infection and coinfection in more than half of the participants. In addition, the clinical findings were highly heterogeneous among the children according to the assay result.