Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Colombiana de Entomología
Print version ISSN 0120-0488On-line version ISSN 2665-4385
Rev. Colomb. Entomol. vol.35 no.2 Bogotá July/Dec. 2009
Molecular characterization of a Colombian Bacillus thuringiensis strain with activity against Tecia solanivora (Lepidoptera: Gelechiidae)
Caracterización molecular de una cepa colombiana de Bacillus thuringiensis con actividad contra Tecia solanivora (Lepidoptera: Gelechiidae)
DIEGO VILLANUEVA1, NUBIA VELÁSQUEZ2, ESPERANZA RODRÍGUEZ3, SERGIO ORDUZ4 and RAFAEL ARANGO5
1 Biologist M. Sc. Grupo de Biotecnología Vegetal, Corporación para Investigaciones Biológicas. Medellín, Colombia.dvillanueva@cib.org.co.
2 Microbiologist MSc. Unidad de Biotecnología Vegetal, Corporación para Investigaciones Biológicas. Medellín, Colombia. nubiayin@gmail.com.
3 Biologist M. Sc., PhD Candidate. Unidad de Biotecnología Vegetal, Corporación para Investigaciones Biológicas. Medellín, Colombia. erodriguez@cib.org.co.
4 Biologist Ph. D. Unidad de Biotecnología y Control Biológico, Corporación para Investigaciones Biológicas. Medellín, Colombia. Escuela de Biociencias, Facultad de Ciencias, Universidad Nacional de Colombia, Sede Medellín, Colombia. sorduz@cib.org.co.
5 M. D. Ph. D. Unidad de Biotecnología Vegetal, Corporación para Investigaciones Biológicas. Medellín, Colombia. Escuela de Biociencias, Facultad de Ciencias, Universidad Nacional de Colombia, Sede Medellín, Colombia. Cra. 72A # 78B-141. Medellín, Colombia. rarango@cib.org.co. Autor para correspondencia.
Received: 5-apr-09 - Accepted 26-sep-09
Abstract: The Guatemalan potato moth, Tecia solanivora (Lepidoptera: Gelechiidae), is one of the most important pests affecting potatoes in northern South America, causing crop losses ranging from 50 to 100%. In this work, we isolated a native strain of Bacillus thuringiensis (146-15801) with high activity against this moth, and we characterized its Cry genes. This strain was the most active out of eleven strains tested against T. solanivora in laboratory bioassays. Specific PCR and genome walk allowed the isolation of Cry genes showing 99 and 95% similarity to the sequences reported in Cry1Ac and Cry2 genes, respectively. Immunolocation experiments demonstrated that Cry1Ac and Cry2 bind to the midgut epithelial cells, suggest that these two proteins are involved in the insecticidal activity against T. solanivora. Results obtained in this work are relevant because native strains comprise a source of biological components for the development of new bioinsecticides, as well as new Cry genes that can be used for biological control.
Key words: Delta-endotoxins. Genome walk. Sequence analysis.
Resumen: La Polilla Guatemalteca de la Papa, Tecia solanivora (Lepidoptera: Gelechiidae), es una de las plagas más importantes del cultivo de la papa en el norte de Sur América, causando pérdidas que oscilan entre el 50 y el 100% de la cosecha. En este trabajo aislamos una cepa nativa de Bacillus thuringiensis (146-15801) con alta actividad contra esta polilla y caracterizamos sus genes Cry. Esta cepa fue la más activa de once cepas evaluadas contra T. solanivora en bioensayos en el laboratorio. Técnicas de PCR específica y caminado genómico permitieron el aislamiento de los genes Cry mostrando un 99 y 95% de similitud con los genes reportados para Cry1Ac y Cry2, respectivemente. Experimentos de inmunolocalización demostraron que Cry1Ac y Cry2 se unen a las células epiteliales del intestino medio del insecto, sugiriendo que estas proteínas están involucradas en la actividad insecticida contra T. solanivora. Los resultados obtenidos en este trabajo son relevantes debido a que las cepas nativas constituyen una fuente de componentes biológicos para el desarrollo de nuevos bioinsecticidas, así como nuevos genes Cry que puedan ser usados para el control biológico.
Palabras clave: Delta-endotoxinas. Caminado genómico. Análisis de secuencia.
Introduction
The Guatemalan Potato Moth (GPM) (Tecia solanivora Povolny, 1973) (Lepidoptera: Gelechiidae) was first described in Central America (Povolny 1973) and is ccurrently the most important entomological pest affecting potato (Solanum tuberosum L.) in Central America and northern South America (Valderrama et al. 2007). Its larvae attack tubers, both in the field and in storage causing losses that range from 50 to 100% (Zeddam et al. 2008)
Pest control is usually made by intensive use of chemical insecticides, potentially generating insect resistance and environmentally associated problems (MacLeod 2005; Valderrama et al. 2007). Bacillus thuringiensis Berliner, 1915 is either a useful alternative or complementary approach to synthetic chemical pesticide applications in commercial agriculture. During sporulation, B. thuringiensis cells produce proteina-ceous Crystalline inclusions composed of δ-endotoxins which specifically kill insect larvae. The specificity of these toxins is determined by their binding affinity to the midgut receptors of the apical membrane of brush border epithelial cells. This is followed by insertion into the cell membrane to form pores, causing paralysis and death (Grochulski et al. 1995; De Maagd et al. 1999). Each type of Cry toxin has a unique spectrum of activity and targets only a small range of insect species. Within this small host target range, there are great differences in potency between species that are often closely related (Gilliland et al. 2002).
In order to have the best Cry proteins for insect control and to prevent or reduce the appearance of insect resistance to current available δ-endotoxins, it is important to identify new B. thuringiensis strains containing Cry proteins with high toxicity against T. solanivora. Although, it has been reported that Cry1Ac toxin from B. thuringiensis subsp. kurstaki is active against this insect (Valderrama et al. 2007), it is unknown which toxic protein is more specific and which has better activity against T. solanivora.
In this study we report the identification of a native B. thuringiensis strain isolated in Colombia, with high toxic activity against T. solanivora. The strain was characterized by sequence analysis of its Cry genes and the immunolocation of the Cry proteins in the intoxicated insect. The availability of this strain and genes provides new alternatives for the control of this insect and their use as control agents for other pests.
Materials and Methods
Tecia solanivora larvae. Tecia solanovora (Lepidoptera: Gelechiidae) larvae were obtained from a colony maintained in the laboratory of Plant Biotechnology of the Corporación para Investigaciones Biológicas, (CIB) (Londoño & Montoya 1994).
Strains, culture conditions and protoxin production. Eleven B. thuringiensis strains isolated in Colombia, with previously known genotypes, were initially used in this study (Table 1). The B. thuringiensis serovar kurstaki strain HD-1 was used as positive control. The strains were supplied by the Biotechnology and Biological Control Unit of CIB.
Strains were grown in M1 medium (Orduz et al. 1992) at 30ºC for 48 h. To solubilize the protoxins this culture was centrifuged at 6000 rpm in a Sorvall-Kendro centrifuge (Ashville, United States), for 10 min, the pellet was resuspended in 50 mM NaOH, 10 mM EDTA solution and incubated at 30ºC for 2 h. This solution was centrifuged at 6000 rpm for 15 min at 4ºC and the supernatant was collected. Total protein concentration of the solubilized protoxins was determined by Bradford assay (Bradford 1976) using bovine serum albumin as standard.
Bioassays. In order to determine the 50% lethal concentration (LC50), the solubilized protoxins were evaluated using six concentrations starting from 300 ng/ul to 9.4 ng/ul with a 0.6 dilution factor in PBS buffer (Sambrook & Russell 2001). Five replicates of cylindrical pieces of potato (Solanum tuberosum var. capira) per concentration were immersed once in the solutions for 5 min and allowed to dry at room temperature (25 ± 2°C). These pieces were placed in disposable covered 5 oz plastic cups and each potato piece was infested with five first-instar larvae and left at a constant temperature (23 ± 1°C) and humidity (75 ± 1%) during eight days. After this period, the number of dead larvae was scored, statistically significant differences were identified by Dunnett Test and the LC50s of the toxins were estimated by Probit analysis on at least three independent bioassays (Dunnett 1955; Finney 1971).
PCR amplification of Cry genes. The most active B. thuringiensis native strain (coded 146-15.801) was cultured overnight at 30°C and shaked at 220 rpm in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl; pH 7.0). 1 ml of culture was centrifuged and the pellet was resuspended in 100 μl of distilled water, boiled for 10 min, and spun at 13.000 rpm for 5 min. The supernatant was collected and used as a source of DNA template for PCR amplification using a series of primers based on conserved regions of Cry1Ac genes (primers Cry001 to Cry004, Table 1).
For amplification of Cry2 gene, previously described UN2 primers were used (Ben-Dov et al. 1997). PCR reactions contained 1 μl of template DNA, 1X reaction buffer (50 mM KCl, 10 mM Tris-HCl pH 8.0), 1.5 mM MgCl2, 0.2 mM dNTPs, 0.4 μM primer and 1.5 U of Taq DNA polymerase (MBI Fermentas, Vilnius, Lithuania). The amplifications were carried out in a iCycler termocycler (Bio-Rad, CA, USA), with an initial step of 94°C for 3 min, followed by 25 cycles of 92ºC for 1min; 51ºC for 30 s and 72ºC for 1 min, and an extra cycle with an extension of 72ºC for 10 min. The PCR products were analyzed by electrophoresis on a 1% agarose gel supplemented with 0.1% ethidium bromide and visualized in a UV transilluminator (Fotodyne Inc., Harhand, WI, USA).
Sequencing of Cry genes. The PCR products were cloned into pGEM-T Easy Vector System I (Promega, Madison, WI, USA), and sequenced using an ABI 3730 DNA sequencing machine as a service provided by Macrogen (Seoul, Korea). In order to obtain the complete sequence of Cry genes, cloning of flanking DNA regions was achieved by genome walking (Siebert et al. 1995). Briefly, 5 μg of the most active B. thuringiensis native strain DNA was digested with four restriction enzymes (SmaI, EcoRV, PvuII and RsaI) and ligated with genome walker adaptors. The ligated fragments were used for an initial PCR amplification with an outer adaptor- specific primer (AP1) and outer gene-specific primers (12GW1, 12GW2 and Cry2GW1, Cry2GW2) (Table 2). The products of these reactions were used as template for a second PCR amplification with the nested adaptor primer AP2 and nested gene specific primers (12GW3, 12GW4 and Cry2GW3, Cry2GW4) (Table 2). This method produced PCR fragments that extended from the known sequence of Cry genes into adjacent DNA, both 5´ and 3´ of the known sequence depending on the location and sequence of gene specific primers. The PCR products were analyzed by 1% agarose gel electrophoresis. The PCR purified products were ligated into pGEM-T Easy Vector System I (Promega) and sequenced. The complete sequences of the encoding region of each gene were confirmed by three independent PCR reactions using high fidelity Pfu polymerase (MBI Fermentas).
Sequence analysis. Sequences obtained of Cry1Ac and Cry2 genes from 146-15.801 strain were manually edited using BIOEDIT Sequence Alignment Editor v7.0.5.2 (Hall 1999), and a multiple alignment was made with the reported sequence of Cry genes in the NCBI/GenBank. Both nucleotide and amino acid sequence differences were obtained using ClustalW (Hall 1999).
Immunolocation. An inmunolocation assay was performed with commercial anti Cry1Ac, anti Cry1Ab and anti Cry2A antibodies (Abraxis, Warminste, PA) in order to study the binding of the Cry toxins of the native Bacillus thuringiensis strains to the cell membrane of midgut cells of first instar T. solanivora larvae. The B. thuringiensis serovar kurstaki strain HD-1 was used as positive control. Briefly, three days protoxin fed larvae were placed in 100% paraplast blocks and were cut in 5 μm thick slices using a microtome (Leica RM 2135, Suchthilfe Wetzlar, Germany). Slices were incubated in three changes of 100% xylol for 5 min at room temperature and these were dehydrated in an ethanol series of 100%, 85%, 70% and 50% 3 min each at room temperature. Slices were then incubated in 1mg/ml trypsin (Sigma, St Louis, USA) (in 2 N HCl) followed by incubation with 0.1 mM PMSF (Phenyl Methyl Sulfonyl Fluoride) (Sigma, St Louis, USA) for 5 min at room temperature. A treatment with hydrogen peroxide (H2O2) at 0.5% in ethanol, for 30 min followed by another treatment at 6% for 15 min was used to eliminate endogenous peroxidases.
Antibody treatment started with one wash in distilled water for 3 min, followed by three washes with TBS (100 mM Tris-HCl, pH 7.5, 0.9% NaCl and 0,1% Tween 20) 3 min each. Each slice was blocked with 3% BSA (Promega) for 1 h at room temperature followed by incubation of 1 h at 37ºC in a humidity chamber with a 1:1000 dilution of the antibody in TBS (100 mM Tris-HCl, pH 7.5, 0.9% NaCl and 0,1% Tween 20) with 1% BSA. Negative controls were incubated only with TBS. After three washes with TBS, peroxidase conjugated anti-rabbit antibody was used at a dilution of 1:2.500 and incubated for 40 min at 37ºC followed by another three washes with TBS. Development was done with 20 μl of DAB cromogen (DAKO; Denmark) in 1 ml DAB buffer (Imidazole - HCl buffer pH 7.5) for 3 min in the dark at room temperature. For contrast and cell identification each preparation was incubated for 2 s in hematoxylin (Sigma, St Louis, USA) and washed in distilled water. The preparations were let to dry at room temperature and observed under light microscopy.
Results
Toxicity of B. thuringiensis Cry proteins to T. solanivora. Eleven strains were selected for testing based on their Cry genes (Table 2) in order to find B. thuringiensis strains with good activity against T. solanivora. Analysis of variance indicated that the average dead larvae was different between the strains (F = 1.92; α = 0.05; P < 0.05). Dunnett test (Dunnett 1955), showed that these differences greater than 1.25 in absolute value relative to the control were significant. In vivo toxicity tests indicated that there were differences among the strains tested and two of them, 146-16.203 and 146-15.801, presented the highest activity (Table 2). These two strains were more active than the positive control B. thuringiensis subsp. kurstaki. Probit analysis of the strains showed that strain 146-15.801, with a previously known Cry1Ac and Cry2 genotype, had the highest toxic activity with an LC50 of 56.82 ng/ul (Table 2). This strain was selected for the isolation and sequencing of its Cry genes.
Isolation and sequence analysis of Cry1Ac and Cry2 genes. Gene specific PCR amplifications and genome walking allowed the isolation and sequencing of a 3.534 nucleotide fragment from strain 146-15.801 encoding for a Cry1Ac gene; (Bacillus thuringiensis toxin nomenclature committee: Cry- 1Ac24; GenBank Accession EF094884). Comparison of the nucleotide sequence of this gene with reported Cry1Ac genes: GenBank Accessions AF492767, AY225453, AY122057, U89872, M11068, M73249 and AY730621; revealed, as expected, a high homology among them, but also some nucleotide differences (Fig. 1).
The amino acid sequence deduced from Cry1Ac24 gene showed 99% similarity to the reported proteins. However, most Cry1Ac proteins of the GenBank display an isoleucine has an asparagine instead; the reported Cry1Ac proteins show a phenylalanine at position 148, the native Cry1Ac24 protein presents a leucine; the reported Cry1Ac proteins present a proline at position 248, while the 146-15.801 native Cry1Ac24 protein presents a serine; the reported Cry1Ac proteins present an asparagine at position 442, the native Cry1Ac24 protein has no amino acid at this position; and finally at position 507 all proteins exhibit a isoleucine whereas the native Cry1Ac24 protein present a phenylalanine.
In a similar way to the Cry1Ac24 gene, specific PCR gene amplifications and genome walking allowed the isolation and sequencing of a 1902 nucleotides fragment from strain 146-15.801 coding for a 2 gene (Bacillus thuringiensis toxin nomenclature committee: Cry2Aa13; GenBank Accession EF094885). Comparison with the reported Cry2A genes: Gen- Bank Accessions AF200816, M31738, D86064, AF433645, AF441855, M23724, X55416, AF164666, AF336115, AY297091 and X57252, revealed that unlike the Cry1Ac genes, there is a high variability among the Cry2 genes (Fig. 2). Nucleotide differences observed were dispersed throughout the coding region of Cry2 gene. The amino acid sequence deduced from Cry2Aa13 gene showed 94% similarity to reported proteins.
Sequence comparison showed that most of Cry2 proteins in the GenBank present more than 50 amino acid differences throughout the protein, not only with the native protein, but also between Cry2 proteins reported.
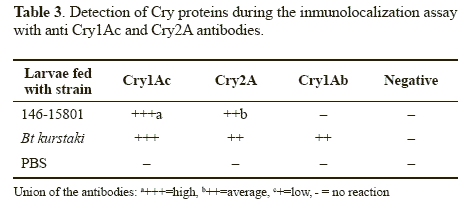
Inmmunolocation. When T. solanivora larvae were fed with 146-15.801 strain or B. thuringiensis subsp. kurstaki strains used as a positive control, there were marked differences in the binding of the toxins to the microvillar brush border of the epithelial cells throughout the midgut of the larvae (Table 3). Cry1Ac24 was the best binding protein followed by Cry- 2Aa13. Figure 3 shows the binding of the different toxins to the midgut epithelial cells.
Discussion
In this work we looked for the implementation of an efficient control mechanism for Tecia solanivora by identifying a native strain of B. thuringiensis with high activity, because few studies have reported which toxin from B. thuringiensis have potential activity against this moth (Valderrama et al. 2007). We found a Colombian strain with high activity against Tecia solanivora. Genes coding for δ-endotoxins were isolated and characterized. Differences in their nucleotide and amino acid sequences with Cry1Ac and Cry2A genes reported were determined. Furthermore we studied the binding of the different B. thuringiensis toxins to the midgut of the insect by inmunolocation.
Eleven strains tested in this study showed activity against T. solanivora. Nevertheless, strains 146-16.203 (Cry1Aa, Cry2) and 146-15801 (Cry1Ac, Cry2) showed the highest activity against the insect. Amongst the two strains selected, 146-15.801 strain showed the lowest LC50 and for this reason was chosen as a source to isolate and to study its Cry1Ac and Cry2 δ-endotoxin genes. The Cry1Ac and Cry2 genes of the most active strains are consistent with previous reports showing that Cry1Ac toxin has good activity against T. solanivora (Valderrama et al. 2007).
Multiple sequence alignment between the 3.534 nucleotides of 146-15.801 Cry1Ac24 gene and other reported Cry1Ac genes showed a 99% homology. Nevertheless, small differences were found located in specific zones previously described to have variability in these genes (Crickmore et al. 1998; Schnepf et al. 1998). Multiple sequence alignment showed a much higher variability spread in Cry2Aa13 gene when compared to Cry1Ac24 sequence.
Cry1 proteins have five conserved blocks located in three functional domains: domain I is involved in the pore formation of the epithelial cell membrane; domain II is involved in receptor binding; and domain III is related to the resistance or susceptibility of the toxin to the proteases of the insect, giving stability to the structure and involved in receptor binding (Peferoen 1997; Crickmore et al. 1998; Schnepf et al. 1998). The amino acid variations found in the Cry1Ac native protein were located in the domain I (L148; S248), domain II (absence of N442) and domain III (F507). Although it is not possible to say that these variations of Cry1Ac24 native protein affect the functionality of the toxin, these could be responsible for the high activity of the strain against T. solanivora. Several studies have shown that the functionality of each domain varies selectively against different insects when point mutations occur (Peferoen 1997; Crickmore et al. 1998; Schnepf et al. 1998).
Concerning Cry2Aa13 protein, when it was compared to Cry2A proteins published, we found 45 amino acids that were different. This high variability could be related to a higher host range or to the fact that it is involved in dual activity against lepidopterans and dipterans (Peferoen 1997; Crick-more et al. 1998; Schnepf et al. 1998). Immunolocation experiments showed that both Cry1Ac24 and Cry2Aa13, bind to the surface of midgut cells of T. solanivora larvae. The specific function of these proteins against this insect must be examined further but it could be possible that both proteins contribute to the toxicity. Combination of Cry1Ac and Cry2Aa genes could be a good alternative to control this insect pest and might to reduce the development of insect resistance against either Cry1Ac24 or Cry2Aa13 proteins.
Conclusions
Two native genes coding for Cry1Ac24 and Cry2Aa13 proteins have been isolated and characterized from a native B. thuringiensis strain with high activity against one of the most important potato pests in northern South America (T. solanivora). Some changes in amino acids could be responsible for the high activity against T. solanivora. Isolation and sequence analysis of these new Cry genes increase the development of the use of Cry proteins as competitive biological insecticides. New isolation of B. thuringiensis in the future in conjunction with knowledge of their toxins will be essential in order to find optimal alternatives to control this insect, diminishing in this way problems associated with chemical insecticides.
Acknowledgements
This work was supported in part by the Instituto Colombiano para el Desarrollo de la Ciencia y la Tecnología “Francisco Jose de Caldas” (Colciencias), Colombia grant 2213-12-13769, CEVIPAPA, Fondo Hortofrutícola Grant CV-03-011-05, the Corporación para Investigaciones Biológicas, CIB and the Postgraduate program in Biotechnology of the Universidad Nacional de Colombia, sede Medellín. We thank Dr. Fernando Ángel Sanchez for critical reading of the manuscript.
Literature cited
BEN-DOV, E.; ZARITSKY, A.; DAHAN, E.; BARAK, Z.; SINAI, R.; MANASHEROB, R.; KHAMRAEV, A.; TROITSKAYA, E.; DUBITSKY, A.; BEREZINA, N.; MARGALITH, Y. 1997. Extended screening by PCR for seven Cry-group genes from field- collected strains of Bacillus thuringiensis. Applied and Environmental Microbiology 63 (12): 4883-4890. [ Links ]
BRADFORD, M. M. 1976. A rapid and sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72: 248-254. [ Links ]
CRICKMORE, N.; ZEIGLER, D. R.; FEITELSON, J.; SCHNEPF, E.; VAN RIE, J.; LERECLUS, D.; BAUM, J.; DEAN, D. H. 1998. Revision of the nomenclature for the Bacillus thuringiensis pesticidal Crystal proteins. Microbiology and Molecular Biology Reviews 62 (3): 807-813. [ Links ]
DE MAAGD, R. A.; DIRK, B.; WILLEM, S. 1999. Bacillus thuringiensis toxin-mediated insect resistance in plants. Trends in Plant Science 4 (1): 9-13. [ Links ]
DUNNETT, C. W. 1955. A multiple comparison procedure for comparing several treatments with a control. Journal of the American Statistical Association 50:1096-1121. [ Links ]
FINNEY, D. 1971. Probit analysis. Cambridge University Press (Londres) 400 p. [ Links ]
GILLILAND, A.; CHAMBERS, C.; BONE, E.; ELLAR, D. 2002. Role of Bacillus thuringiensis Cry1 δ-endotoxin binding in determining potency during Lepidopteran larval development. Applied and Environmental Microbiology 68: 1509-1515. [ Links ]
GROCHULSKI, P.; MASSON, L.; BORISOVA, S.; PUSZTAICAREY, M.; SCHWARTZ, J. L.; BROUSSEAU, R.; CYGLER, M. 1995. Bacillus thuringiensis CryIA(a) insecticidal toxin: Crystal structure and channel formation. Journal of Molecular Biology 254 (3): 447-464. [ Links ]
HALL, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95-98. [ Links ]
LONDOÑO, M.; MONTOYA, O. 1994. Cría de Tecia solanivora Povolny para ensayos biológicos. ICA-CORPOICA (Colombia). 35 p. [ Links ]
MacLEOD, A. 2005. Data sheets on quarantine pests: Tecia solanivora. European and Mediterranean Plant Protection Organization 35: 399-401. [ Links ]
ORDUZ, S.; ROJAS, W.; CORREA, M. M.; MONTOYA, A. E.; DE BARJAC, H. 1992. A new serotype of Bacillus thuringiensis from Colombia toxic to mosquito larvae. Journal of Invertebrate Pathology 59: 99-103. [ Links ]
PEFEROEN, M. 1997. Progress and prospects for field use of Bt genes in crops. Trends in Biotechnology 15 (5): 173-177. [ Links ]
POVOLNY, D. 1973. Scrobipalpopsis solanivora sp. n. -A new pest of potato (Solanum tuberosum) from Central America. Acta Universitatis Agriculturae, Facultas Agronomica 21: 143-146. [ Links ]
SAMBROOK, J.; RUSSELL, D. 2001. Molecular cloning. A laboratory manual. 3a ed. Cold Spring Harbor Laboratory Press. New York. 1252 p. [ Links ]
SCHNEPF, E.; CRICKMORE, N.; VAN RIE, J.; LERECLUS, D.; BAUM, J.; FEITELSON, J.; ZEIGLER, D.; DEAN, D. 1998. Bacillus thuringiensis and its pesticidal Crystal proteins. Microbiology and Molecular Biology Reviews 62 (3): 775-806. [ Links ]
SIEBERT, P.; CHENCHIK, A.; KELLOGG, D.; LUKYANOV, K.; LUKYANOV, S. 1995. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Research 23 (6): 1087-1088. [ Links ]
VALDERRAMA, A.; VELASQUEZ, N.; RODRIGUEZ, E.; ZAPATA, A.; ZAIDI, M.; ALTOSAAR, I.; ARANGO, R. 2007. Resistance to Tecia solanivora (Lepidoptera: Gelechiidae) in three transgenic andean varieties of Potato expressing Bacillus thuringiensis Cry1Ac protein. Journal of Economic Entomology 100 (1): 172-179. [ Links ]
ZEDDAM, J.; ORBE, K.; LÉRY, X.; DANGLES, O.; DUPAS, S.; SILVAIN, J. 2008. An isometric virus of the potato tuber moth Tecia solanivora (Povolny) (Lepidoptera: Gelechiidae) has a trisegmented RNA genome. Journal of Invertebrate Pathology 99 (2): 204-211. [ Links ]