Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Colombiana de Entomología
Print version ISSN 0120-0488
Rev. Colomb. Entomol. vol.38 no.1 Bogotá Jan./June 2012
Biotype characterization and genetic diversity of the greenbug, Schizaphis graminum (Hemiptera: Aphididae), in north Tunisia
Caracterización de biotipo y diversidad genética del pulgón verde, Schizaphis graminum (Hemiptera: Aphididae), en el norte de Túnez
IMEN KHARRAT1,2, DHIA BOUKTILA1,3, MAHA MEZGHANI-KHEMAKHEM1,4, HANEM MAKNI1,5 and MOHAMED MAKNI1,6
1 Unit of Research on Genomics of Agricultural Insect Pests (GIRC) - Faculty of Sciences of Tunis - University of Tunis El-Manar (Tunisia).2 Ph. D. student in GIRC.
3 Post-Doc. Associate Professor in the Higher Institute of Biotechnology of Béja - University of Jendouba (Tunisia). Researcher in the GIRC. dhia_bouktila2000@yahoo.fr. Corresponding author.
4 Post-Doc. Associate Professor in the Faculty of Sciences of Tunis - University of Tunis El-Manar (Tunisia). Researcher in the GIRC.
5 Professor in the Higher Institute of Animation for Youth and Culture of Bir-El-Bey - University of Tunis (Tunisia) / Research Director in the GIRC.
6 Professor in the Faculty of Sciences of Tunis - University of Tunis El-Manar (Tunisia) / Research Director in the GIRC.
Received: 20-oct-2011 - Accepted: 24-may-2012
Abstract: The greenbug Schizaphis graminum, is a major pest of wheat worldwide. Biotype screening of this pest is essential to develop pest management programs. In this research, eight greenbug clones, collected on wheat in the cereal-growing region of Béja (north Tunisia), were used to determine their damage on six reference wheat cultivars. All tested clones shared a unique biotypic profile, similar to biotype C. Moreover, DNA from the tested clones and that from seven reference clones of biotypes C, E, F, G, H, I and K, was analyzed, using 5 RAPD-PCR primers. The UPGMA method clustered samples into two distinct clades: a first one (I) included clones from north Tunisia, which were clearly associated to agricultural biotypes C, E, I and K, while a second clade (II) included non agricultural biotypes F, G and H. Results reported in this paper suggest that resistance genes Gb2, Gb3, Gb4, Gb5 and Gb6 in wheat would be the most efficient if used in wheat improvement programs for resistance against greenbug in Tunisia.
Key words: Aphid. Cereals. Resistance genes. Insect genotypes. Molecular markers.
Resumen: El pulgón verde Schizaphis graminum, es una plaga importante del trigo en todo el mundo. La detección de biotipos de esta plaga es esencial para el desarrollo de programas de manejo de plagas. En esta investigación se colectaron, ocho clones del pulgón verde, en el trigo en la región de cultivo de cereales de Béja (norte de Túnez), y se utilizaron para determinar el daño en seis cultivos de trigo de referencia. Todos los clones ensayados comparten un perfil único de biotipo, similar al biotipo C. Además, el ADN de los clones ensayados y de los siete clones de referencia de biotipos C, E, F, G, H, I y K, se analizaron, utilizando 5 RAPD PCR primers. El método UPGMA agrupó las muestras en dos clados: el primero (I) incluye clones del norte de Túnez, que están claramente asociados a los biotipos agrícolas C, E, I y K, mientras que un segundo clado (II) incluyó los biotipos no agrícolas F, G y H. Los resultados sugieren que los genes de resistencia, la Gb2, Gb3, Gb4, Gb5 y Gb6 en el trigo serían los más eficaces si se utilizan en programas de mejoramiento de trigo para la resistencia contra el pulgón verde en Túnez.
Palabras clave: Pulgón. Cereales. Genes de resistencia. Genotipos de insectos. Marcadores moleculares.
Introduction
The greenbug, Schizaphis graminum Rondani (Homoptera: Aphididae) is an aphid pest of several graminaceous crops worldwide. It causes severe injuries to host plants in all growth stages and often kills the entire cereal plant. It is prejudicial to the host, either due to the large quantity of sap it extracts, causing water and nutrients depletion (Cruz et al. 1998) or due to vectoring Barley Yellow Dwarf Virus (BYDV, Gray et al. 2007), Mosaic Maize Dwarf Virus (MMDV, Nault and Bradley 1969) or Sugarcane Mosaic Virus (SCMV, Ingram and Summers 1938). Over twenty greenbug biotypes have been recognized (Porter et al. 1997; Shufran et al. 2000; Burd and Porter 2006); nearly all of them (except D, which has been identified on the basis of insecticide resistance) have been characterized based on the preference for a host plant species and/or ability to damage specific cultivars of a defined species.
The term "biotype" usually designates an infraspecific group of organisms that are not morphologically distinguishable, but differing by a biological function (Eastop 1973).
Within this definition, S. graminum biotypes could be described as a case of host race. In fact, host plant response remains the main criterion for recognition of greenbug biotypes. However, determination of host plant response is often laborious and time-consuming. Therefore, other methods based on morphological characters (Starks and Burton 1977), isozymes (Abid et al. 1989) and mitochondrial DNA (Shufran et al. 2000), have been used to assess genetic relationships among biotypes or to develop alternative identification procedures. These methods, however, have not fully distinguished all biotypes. Random Amplified Polymorphic DNA-Polymerase Chain Reaction (RAPD-PCR) has been successfully applied to reveal distinctive patterns among some greenbug biotypes (Black et al. 1992; Aikhionbare et al. 1998; Lopes-Da-Silva et al. 2004). Because biotype identification of S. graminum should be the first step for any cereal-breeding program aiming to obtain resistant cultivars to this pest, the main objective of this work was to characterize the biotype(s) of this insect in a wheat-growing area in the north of Tunisia.
Materials and Methods
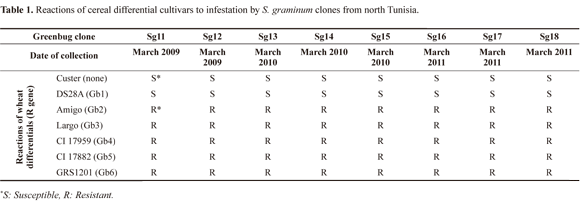
Insect and plant material. Eight greenbug clones (Sg11-Sg18), collected over a three year-period, in March 2009, 2010 and 2011 (Table 1), from several wheat fields located in Beja, in the north of Tunisia (36°43'N 9°11'E), were used for infestation assays and subsequent molecular analysis. Seven clones of biotypes C, E, F, G, H, I and K, received from the USDA-ARS, Stillwater, Oklahoma (USA), were used in molecular analysis. All clones were reared for 3 generations on the susceptible barley cultivar 'Custer', in a growth chamber, under standard conditions (22-5°C, 50% relative humidity and 16L: 8D photoperiod, Shufran et al. 1992). In addition, 6 differential wheat cultivars ('DS28A', 'Amigo', 'Largo', 'CI 17959', 'CI 17882', and 'GRS1201', Burd and Porter 2006), containing each a resistance gene to S. graminum were used in monitoring greenbug biotype. A barley cultivar ('Custer'), containing no resistance gene was used as susceptible check. All these cultivars (Table 1) were kindly provided by USDA-ARS, Stillwater, Oklahoma, USA.
Determination of biotype. The biotype for greenbug clones from Tunisia was determined by screening the feeding damage on the wheat differentials mentioned in table 1. As described in Starks and Burton (1977), 10 seeds of each wheat differential and 10 seeds of the susceptible check 'Custer' were planted in 15cm rows (replicated four times) in flats on greenhouse benches. Seedlings were grown at 25°C with a 16:8 (L:D) photoperiod. Flats were caged to avoid secondary infestation. Immediately after emergence, plants were infested by greenbug clones. Once the susceptible control plants (Custer) were killed (usually within 7 - 14 d), the test was terminated and the plants were scored as alive (resistant) or dead (susceptible).
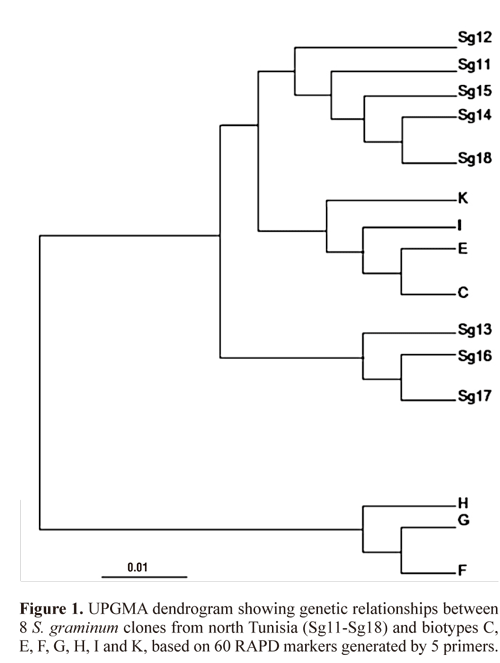
RAPD-PCR analysis. Following the determination of biotype, Tunisian greenbug clones were freshly collected from plants and kept in Eppendorf® tubes; while other clones, belonging to selected biotypes, were stored in ethanol 95%. DNA was isolated, from all clones, according to the protocol described in Doyle and Doyle (1987). RAPD-PCR reactions were performed in a Thermal cycler 2720 (Applied Biosystem, USA), as described in Lopes-Da-Silva et al. (2004). Five single decamers (Operon Technologies, USA) were used as primers; OPC-09 (5'-CTCACCGTCC-3'), OPG-03 (5'- GAGCCCTCCA-3'), OPG-08 (5'-TCACGTCCAC-3'), OPG-18 (5'- GGCTCATGTG-3') and OPH-05 (5'- AGTC-GTCCCC-3'). Amplification products were analyzed on 1.5% agarose gel in Tris-borate-EDTA buffer, stained in ethidium bromide and visualized under UV light. A 100 bp ladder (Invitrogen) was used as molecular size standard. The RAPD-PCR patterns, for each clone, were identified visually by scoring the presence or absence of all reproducible bands and a single binomial genotype, for each of the 15 samples, was defined. A RAPD fragment was considered as polymorphic once it was present in at least one S. graminum clone and absent in the remaining clones. The percentage of polymorphic fragments (%P) was calculated as (number of polymorphic fragments / total number of fragments generated) x 100. Pairwise genetic distances (Nei and Li 1979) between clone pairs were calculated using the GENDIST program of PHY-LIP software version 3.68 (Felsenstein 2008). Finally, a UP-GMA dendrogram was constructed on the basis of pairwise Nei and Li's (1979) genetic distances, using the NEIGHBOR program of PHYLIP version 3.68 (Felsenstein 2008) and visualized by the software Treeview version 1.6.6 (Page 1996).
Results and Discussion
Biotype screening on a differential set of indicator cultivars showed that the studied greenbug clones from northern Tunisia exhibited profile similar to biotype C (Table 1). All DNA amplification products obtained were reproducible and 60 different markers were scored with the 5 primers used, ranging from 150 to 1100 bp in size. The percentage of polymorphic RAPD fragments generated by each primer alone varied from 60% with primer OP-C09 to 88.88% with primer OP-G18, with an overall percentage of 74.05%. The range of genetic distance was high, varying from 0.040 between clones Sg11 and Sg15, to 1.021 between biotype E and clone Sg11. The dendrogram yielded by the UPGMA method (Fig. 1) showed that the studied clones clustered into 2 distinct clades: a first clade (I), included S. graminum clones from north Tunisia, which were clearly associated to agricultural biotypes C, E, I and K (Shufran et al. 2000); while a second clade (II) included non agricultural biotypes F, G and H. These results do not contrast with the infestation assay; thereby tending to confirm the assignment of Tunisian clones to biotype C. Although the limited set of primers used in this study did not result in clear discrimination between the reference biotypes, the RAPD analysis was useful to distinguish between agricultural biotypes (C, E, I and K) and non-agricultural ones (F,G and H). In similar studies, Aikhonbare et al. (1998) with USA populations, and Lopes-Da-Silva et al. (2004) with Brazilian populations, could not distinguish biotypes C and E, by RAPD markers.
Although greenbug clones from Tunisia, used in this study, showed a homogeneous behaviour at the physiological level, when used for infesting wheat cultivars; they were heterogeneous in molecular analysis, as they were relatively discarded in the dendrogram generated based on RAPD-PCR. This observation is in agreement with several reports where genetic differences were detectable between and within greenbug biotypes (Black et al. 1992; Shufran et al. 1992; Black 1993; Anstead et al. 2002). While some authors (Black et al. 1993) have treated biotypes as evolutionary lineages/ units, Anstead et al. (2002) suggested that a conceptual distinction should be made between three terms, which are "gentotype", "biotype" and "host race". In other words, a biotype may be characterized by its pattern of virulence to resistance genes or associated with a particular host plant species; yet, it should not be treated as an evolutionary unit or be given taxonomic status and, therefore, it could potentially display genotypic diversity. For example, Shufran et al. (1992) found that biotype E in the field was genetically heterogeneous and concluded that it was comprised of many clonal lineages. In order to distinguish normal variation "between clones within a biotype" from differences "between biotypes", Anstead et al. (2002) suggest that phylogeographical surveys should include large samples and integrate data from different genomic and mitochondrial markers. Anyway, biotype designation remains very useful to researchers, and especially to breeders, as it enables the identification of efficient resistant genes. Biotype C, characterized in this study, implies that resistance genes Gb2, Gb3, Gb4, Gb5 and Gb6 in wheat would be efficient if integrated in wheat improvement programs in Tunisia. The transfer of these genes into commercial wheat cultivars in Tunisia could be facilitated by a marker-assisted selection (MAS), as several molecular markers associated with most genes have been characterized (Weng et al. 2005; Lu et al. 2010).
For a better efficiency of cereal breeding programs, the findings reported here should be continued by extensive surveys, to identify greenbug biotypes in Tunisia, at a greater scale encompassing the South of the country where barley and sorghum are usually grown. Besides, regular biotype surveys, through field and molecular assays, should be conducted, as biotype shifts may occur, rendering previously efficient genes, susceptible to the new biotypes.
Acknowledgements
The authors thank the USDA-ARS (Stillwater, Oklahoma-USA) for providing plant and insect material. Special thanks are addressed to the referees who have reviewed this work for "Revista Colombiana de Entomologia". This work was funded by a "PRF" project between the Ministry of Higher Education (Tunisia) and the Ministry of Agriculture (Tunisia).
Literature cited
ABID, H. S.; KINDLER, S. D.; JENSEN, S. G.; THOMAS-COMPTON, M. A.; SPOMER, S.M. 1989. Isozyme characterization of sorghum aphid species and greenbug biotypes (Homoptera: Aphididae). Annals of the Entomological Society of America 82: 303-306. [ Links ]
AIKHIONBARE, F. O.; PRUESS, K. P.; MAYO, Z. B. 1998. Greenbug (Homoptera: Aphididae) biotypes characterized using random amplified polymorphic DNA Genetic Analysis. Biomolecular Engineering 14: 105-108. [ Links ]
ANSTEAD, J. A., BURD, J. D., SHUFRAN, K. A. 2002. Mitochondrial DNA sequence divergence among Schizaphis graminum (Hemiptera: Aphididae) clones from cultivated and non-cultivated hosts: haplotype and host associations. Bulletin of Entomological Research 92: 17-24. [ Links ]
BLACK, W. C.; DUTEAU, N. M.; PUTERKA, G. J.; NECHOLS,J. R.; PETTORINI, J. M. 1992. Use of Random Amplified Polymorphic DNA-Polymerase Chain Reaction (RAPD-PCR) to detect DNA polymorphisms in aphids (Homoptera: Aphididae). Bulletin of Entomological Research 82: 151-159. [ Links ]
BLACK, W. C. 1993. Variation in the ribosomal RNA cistron among host-adapted races of an aphid (Schizaphis graminum). Insect Molecular Biology 2: 59-69. [ Links ]
BURD, J.; PORTER, D. 2006. Biotypic diversity in greenbug (Hemiptera: Aphididae): Characterizing new virulence and host associations. Journal of Economic Entomology 99: 959-965. [ Links ]
CRUZ, I.; VENDRAMIN, J. D.; OLIVEIRA, A. C. 1998. Determinacäo do período de avaliacäo de nao preferencia de sorgo ao pulgäo-verde, Schizaphis graminum (Rond.) (Homoptera: Aphididae). Anais Da Sociedade Entomologica Do Brasil 27: 299-302. [ Links ]
DOYLE, J. J.; DOYLE, J. L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Photochemical Bulletin 19: 11-15. [ Links ]
EASTOP, V. F. 1973. Biotypes of aphids. pp. 40-51. In: Lowe, A. D. (Eds.) Perspectives in aphid biology. The Entomological Society of New Zealand Inc. Auckland, New Zealand. [ Links ]
FELSENSTEIN, J. 2008. PHYLIP: Phylogenetic Interference package version 3.68c. Department of Genetics, University of Washington, Seattle, USA. [ Links ]
GRAY, S. M.; CAILLAUD, M. C.; BURROWS, M.; SMITH, D. M. 2007. Transmission of two viruses that cause Barley Yellow Dwarf is controlled by different loci in the aphid, Schizaphis graminum. Journal of Insect Science 25: 1-15. [ Links ]
INGRAM, J. W.; SUMMERS, E. M. 1938. Transmission of sugarcane mosaic by the greenbug (Toxoptera graminum Rond.). Journal of Agricultural Research 56: 537-540. [ Links ]
LOPES-DA-SILVA, M.; TONET, G. E. L.; VIEIRA, L. G. E. 2004. Characterization and genetic relationships among Brazilian biotypes of Schizaphis graminum (Rondani) (Hemiptera: Aphididae), using RAPD markers. Neotropical Entomology 33: 43-49. [ Links ]
LU, H., RUDD, J. C., BURD, J. D., WENG, Y. 2010. Molecular mapping of greenbug resistance gene Gb2 and Gb6 in T1AL.1RS wheat-rye translocations. Plant breeding 129: 472-476. [ Links ]
NAULT, L. R.; BRADLEY, R.H.E. 1969. Acquisition of maize dwarf mosaic virus by the greenbug, Schizaphis graminum. Entomological Society of America 62: 403-406. [ Links ]
NEI, M.; LI, W. H. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the national Academy of Sciences of USA 76: 5269-5273. [ Links ]
PAGE, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Applications in Biosciences 12: 357-358. [ Links ]
PORTER, D. R., BURD, J. D., SHUFRAN, K. A., WEBSTER, J. A., TEETES, G. L. 1997. Greenbug (Homoptera, Aphididae) biotypes selected by resistant cultivars or preadapted opportunists? Journal of Economic Entomology 90: 1055-1065. [ Links ]
SHUFRAN, K. A.; MARGOLIES, A. C.; BLACK, W. C. 1992. Variation between biotype E clones of Schizaphis graminum (Homoptera: Aphididae). Bulletin of Entomological Research 92: 407-416. [ Links ]
SHUFRAN, K. A.; BURD J. D.; ANSTEAD, J. A.; LUSHAI, G. 2000. Mitochondrial DNA sequence divergence among green-bug (Homoptera: Aphididae) biotypes: evidence for host-adapted races. Insect Molecular Biology 9 (2): 179-184. [ Links ]
STARKS, K. J.; BURTON, R. L. 1977. Greenbugs: Determining biotypes, culturing, and screening for plant resistance with notes on rearing parasitoids. USDA Technical Bulletin 1556. U. S. Government Print Office, Washington, D. C. [ Links ]
WENG, Y., LI, W., DEVKOTA, R. N., RUDD, J. C., 2005. Microsatellite markers associated with two Aegilops tauschii-derived greenbug resistance loci in wheat. Theoretical and Applied Genetics 110: 462-469. [ Links ]