Introducción
Whiteflies (Hemiptera: Aleyrodidae) are insects that feed on plants by sucking large quantities of sap. Sucking plant sap can cause early wilting, stunted growth, premature defoliation, and eventually yield loss (Shukla et al. 2016). In addition, whiteflies secrete honeydew that causes sooty mould growth on plants leaves and fruits and reduces their market values. Whiteflies have also been reported to act as vectors for plant viruses (Byrne and Bellows 1991; Jones 2003; Shukla et al.2016). There are around 1556 species of whiteflies in 161 genera (Martin 2004), however, only a few acts as a vector for plant viruses. Bemisia tabaci (Gennadius, 1889) transmits around 212 viruses from five genera Begomovirus, Crinivirus, Ipomovirus, Carlavirus, and Torradovirus (Navas-Castillo et al. 2011; Polston et al. 2014). Trialeurodes vaporariorum (Westwood, 1856), the greenhouse whitefly, transmits viruses from two genera Crinivirus and Torradovirus (Navas-Castillo et al. 2011). Trialeurodes abutiloneus (Haldeman, 1850, the banded-winged whitefly) transmits viruses of the genera Crinivirus and Torradovirus (Mlynarek and Labbé 2018). Bemisia afer (sensu lato) transmits the sweet potato chlorotic stunt virusof Crinivirus (Gamarra et al. 2010; Navas-Castillo et al. 2011). Trialeurodes ricini (Misra, 1924, the castor bean whitefly) could be a vector for the tomato yellow leaf curl virus (TYLCV) in Egypt (Idriss et al. 1997).It is important to study the diversity of the different whitefly populations to be able to manage and design effective control methods for these pests. During the period between 1985-1994, a study was conducted to identify the whitefly species in Jordan and report some of their natural enemies. Eleven species were morphologically identified based on the characteristics of their pupa. These species were Acaudaleyrodes citri (Priesner and Hosny, 1934) Aleurocanthus zizyphi (Priesner and Hosny, 1934), Aleurolobus niloticus (Priesner and Hosny, 1934), Aleurolobus olivinus (Silvestri, 1911), Aleyrodes proletella (Linnaeus, 1758), Aleyrodes singularis (Danzig, 1964), B. tabaci, Siphoninus phillyreae (Haliday, 1835), Trialeurodes lauri (Signoret, 1882), T. ricini (Misra), and T. vaporariorum (Allawi 1994). In addition to the aforementioned whiteflies, Acaudaleyrodes rachipora (Singh, 1931, Babul whitefly) and Aleurolobus marlatti (Quaintance, 1903) have been also reported in Jordan (Ghahari et al. 2009). Nonetheless, the classical taxonomy of whiteflies based on the morphology of the puparium (fourth instar) is complicated since the intraspecific variability in the morphology such as the size, shape, number of setae and papillae, the perianal structure, and the body size, can be affected by the variations in the environment (Ko et al. 2005).The “superbug” Bemisia tabaci is one of the most damag-ing insects known in the agricultural world (Barinaga 1993). It is broadly polyphagous, feeding on an estimated 600 plant species (European and Mediterranean Plant Protection Orga-nization 2004). In Jordan, around 339 plant species in 64 fam-ilies were reported as hosts for B. tabaci (Sharaf and Allawi 1980). B. tabaci was described as a species complex having many putative species that are morphologically indistinguish-able (Dinsdale et al. 2010). Reliable morphological markers which can distinguish between the different genetic groups of the B. tabaci species complex are not known (Rosell et al.1997). As a result, the molecular markers can be an important tool to study the variation in populations of the species com-plex of B. tabaci (Cervera et al. 2000).Phylogenetic studies comparing the sequence of a region of the DNA such as the 16S ribosomal subunit (Frohlich et al.1999), ribosomal internal transcribed spacer-1 ITS1 sequence (De Barro et al. 2000; Wu et al. 2003), and mitochondrial cytochrome oxidase I (mtCOI) gene (Frohlich et al. 1999; Kirk et al. 2000; Luo et al. 2002) have been carried out to determine the genetic relationships among B. tabaci species complex. Dinsdale et al. (2010) performed a study based on the analysis of sequence pairwise divergence and the Bayesian phylogenetic analysis of mtCOI gene to study the B. tabaco species complex. Based on the genetic species concept to distinguish between the putative species, besides the mating experiment, they concluded that B. tabaci is a species complex consisting of 11 groups containing 24 species (Dinsdale et al. 2010). Boykin (2014) concluded that B. tabaci species complex seems to be made up of more than one species. This was based on data obtained from mating compatibility (Xu et al. 2010; Liu et al. 2012; Sun et al. 2011), genomes (Wang et al. 2011; Wang et al. 2013), and mtCOI phylogenetic analysis (Boykin et al. 2007; Dinsdale et al. 2010; Boykin et al. 2012; Tay et al. 2012).This study is concerned with the characterization of the most invasive putative species of B. tabaci, which are the Middle East-Asia Minor 1 (MEAM1; previously described as biotype B), and the Mediterranean putative species (MED; previously known as biotype Q). Both of them are important from a biosecurity perspective, as they are resistant to a wide range of insecticides (Prabhaker et al. 1988), globally inva-sive, and cause huge economic losses (Oliveira et al. 2001; Boykin et al. 2012). MEAM1 invaded at least 54 countries around the world (Broadbent et al. 1989; Cheek and MacDon-ald 1994, De Barro et al. 2011). Whilst MED putative spe-cies invaded at least ten countries worldwide such as United States, China, Japan, and New Zealand (De Barro et al. 2011).Since the putative species MEAM1 and MED of B. tabaci are morphologically indistinguishable (Liu et al. 2016) many studies were performed to understand their dispersal behaviour, insecticide resistance, plant-host preference, endo-symbiont composition, fecundity, and efficiency in plant vi-ruses transmission (Brown et al. 1995a; Horowitz et al. 2005; Bing et al. 2012; Liu et al. 2016; Shi et al. 2018; Watanabe et al. 2019; Yang et al. 2020). MED putative species is char-acterized by its resistance to a wide variety of insecticides (Nauen et al. 2002; Horowitz et al. 2005; Nauen and Denholm 2005; Ghanim and Kontsedalov 2007; Yang et al. 2013). In Jordan, the presence of MEAM1 was documented by (Brown et al. 1995b). Subsequently, a study carried out us-ing RAPD-PCR to identify the B. tabaci species complex in Jordan found MEAM1 and the New World putative species that is formerly known as biotype A (Sharaf and Hasan 2003). The highly invasive putative species MED was not report-ed in Jordan before, although it was reported in neighbour countries such as Syria, Palestine/Israel, and Egypt (Horow-itz et al. 2003; Khasdan et al. 2005; De Barro et al. 2011). This work aims to combine the classical morphological iden-tification method with the DNA barcoding marker (mtCOI) for the first time to identify and document the presence of the whitefly species in Jordan.
Materiales y métodos
Sample collection and morphological identification
To learn more about the status of whiteflies in Jordan, around 111 different whiteflies samples were collected during the years 2009-2012 (Table 1). Of the 111 samples, around 85 were of B. tabaci. The samples of B. tabaci were collect-ed from more than 23 hosts of cultivated plants (e.g. toma-to, cucumber, cauliflower, okra, watermelon, eggplant, and squash), non-cultivated (e.g. basil), and non-food crops (e.g. cotton, poinsettia, and Lantana camara L.). The host plants were grown in both greenhouses and open fields in different geographical regions in Jordan including the Jordan Valley area (32º19’1.20”N 35º34’7.19”E), and another six different provinces: Amman (31º35’1.28”N 36º20’0.06”E), AL-Balqa` (32º00’0.00”N 35º40’0.01”E), Madaba (31º34’59.99”N 35º40’0.01”E), Jerash (32º15’0.00”N 35º55’0.01”E), Ma`an (30º19’59.99”N 36º34’59.99”E), and Al-Mafraq (32º19’59.99”N 37º55’0.01”E). Pupal stages were collected and reared until adults emerged. The collected adults were preserved in 70 % ethanol to be subjected to DNA isolation, whilst the empty pupal cases were sent for morphological identification by Professor Dan Gerling (Tel Aviv University). Two references for the putative species MEAM1 and MED were kindly provided by Dr Rami Horowitz (the Institute of Plant Protection, Gilat Research Centre).
DNA extraction and amplification of the mtCOI gene
DNA was extracted from a single whitefly adult or pupa for DNA barcoding of the collected samples according to (Cenis et al. 1993) with modifications recommended by (Khasdan et al. 2005). After the DNA extraction, a PCR reaction was performed to amplify 816 bp fragment of the mtCOI gene (Khasdan et al. 2005) with some modifications. The PCR reaction contained around 20 ng total DNA in 1X buffer, one unit of the Ta q DNA polymerase and 0.2 μM dNTPs (Promega Corporation, USA), 2.5 mM MgCl2, 0.4 μM of the forward primer C1-J-2195 5`TTGATTTTTTGGT-CATCCAGAAGT3` and 0.4 μM reverse primer L2-N-3014 5`TCCAATGCACTAATCTGCCATATTA3` (Frohlich et al.1999). PCR was performed in a PTC200 thermocycler (MJ Research Inc., USA). The PCR program was composed of an initial denaturation at 94 °C for 3 min, 40 cycles of one min at 94 °C followed by one min at 52 °C and one min at 72 °C, and a final extension step at 72 °C for seven min. The PCR products were analysed on 1 % agarose gel stained with 0.5 μg/ml of ethidium bromide. A part of the amplified mtCOIgene was sent for sequencing and another part was analysed in the following step.
Cleaved Amplified Polymorphic Sequences (CAPS) for (mtCOI) sequences
To reduce the number of the samples that will be sent for sequencing, two diagnostic techniques were used to distinguish between B. tabaci species complex. The first technique was CAPS which distinguishes between MEAM1 and MED putative species that are likely to present in Jordan. To perform CAPS, a part of mtCOI gene, which was amplified in the previous step, was subjected to digestion by restriction endonuclease VspI according to (Khasdan et al.2005). Only a short fragment (41 bp) was cut out of MEAM1, while PCR products of MED yielded three fragments of about 436 bp, 292 bp, and 41 bp.
Bidirectional PCR amplification of mtCOI fragments
The second diagnostic technique was used to distinguish be-tween B. tabaci species complex was the bidirectional PCR (Tsagkarakou et al. 2007). Four primers were used in each PCR reaction. The two outer primers, the forward primer C1-J-2195 (5` TTGATTTTTTGGTCATCCAGAAGT 3`; Frohlich et al. 1999) and the reverse primer tRNA-1576 (5` TATAAATCTTAAATTTACTGCA 3`; Tsagkarakou et al.2007) they yielded around 879 bp control fragment for all the B. tabaci species complex. The two inner primers, LQ 5` AAGGGGCCTGAATTTATTG 3` and RB5` CTACTTTGG-GTGGAATAAAGTCT 3` were designed and tested to distin-guish between MEAM1 and MED (Tsagkarakou et al. 2007). In the case of MEAM1 putative species, RB/tRNA1576 primers amplify the 609 bp fragment. Whilst, in the case of MED LQ/C1-J-2195 primers will amplify the 310 bp fragment. If only the control band is obtained, it may indicate that it belongs to another putative species of the B. tabaci such as the New World or other putative species which were known pre-viously as C, E, and G biotypes and the exact biotype will be confirmed by the sequencing of the mtCOI gene. The PCR reaction was performed in mostly the same way as above, with the exception that in this reaction, four primers were used and the PCR program was, initial denaturation at 94 °C for 3 min, 40 cycles of 45 sec at 94 °C, one min at 50 °C and one min at 72 °C, and a final extension step at 72 °C for 10 min.
Sequencing and phylogenetic analysis of mtCOI gene
Representative samples of B. tabaci species complex and the other whitefly species were sent for sequencing at Mac-rogen, (Seoul, South Korea). Analysis of the sequences was performed using MEGA X (Kumar et al. 2018) and the Nu-cleotide Basic Local Alignment Search Tool (Nucleotide BLAST) service provided by the National Center for Bio-technology Information (NCBI) (Zhang et al. 2000; Morgu-lis et al. 2008). The obtained sequences were submitted to GenBank. To illustrate the relationship between the sequences of the whiteflies obtained from Jordan and other whiteflies sequences from all over the world, a phylogenetic tree was constructed. The sequences of whiteflies from Jordan were aligned and trimmed to the same length beside other sequenc-es of whiteflies from throughout the world obtained from the GenBank. Then the phylogenetic tree was built using Max-imum Likelihood method based on the Tamura-Nei model (Tamura and Nei 1993).
Resultados
Ten species of whiteflies were morphologically identified in Jordan
As the aim of this study is to combine the morphological and molecular identification tools to identify the whitefly species in Jordan; the morphological identification was the main tool for identifying the whitefly species other than B. tabaci. According to the morphological classification of the whitefly species, nine different whitefly species were identified in addition to B. tabaci (Table 1). The identified species were Trialeurodes lauri (Signoret), Trialeurodes ricini (Misra), Aleyrodes singularis (Danzig), Aleurolobus niloticus (Priesner and Hosny), Aleurolobus olivinus (Silvestri), Acaudaleyrodes rachipora (Singh), Africaleurodes coffeacola (Dozier, 1934), Siphoninus phillyreae (Haliday) and Tetraleurodes neemani (Bink-Moenen, 1992). They belong to seven different genera. The two whitefly species Africaleurodes coffeacola and T. neemani had not been recorded before in Jordan.
Table 1 Updated whitefly species in Jordan, host plants, location and method(s) used in species identification
| Whitefly species | Host | Location | Accession No. (GenBank) | Identification method |
|---|---|---|---|---|
| Acaudaleyrodes rachipora | Citrus limon (L.) | Amman | KP418775 | Morphology and molecular |
| Osbeck Olea europaea L. | KP418776 | |||
| KP418780 | ||||
| Africaleurodes coffeacola | Ziziphus spina-christi (L.) Desf | Al-Balqa` | Morphology | |
| Aleurolobus niloticus | Punica granatum L. | Al-Balqa` | KP418772 | Morphology and molecular |
| KP418773 | ||||
| Aleurolobus olivinus | Olea europaea L. | Amman, Al-Balqa` | KP418774 | Morphology and molecular |
| KP418778 | ||||
| KP418779 | ||||
| Aleyrodes singularis | Lactuca serriola L. | Amman, AL-Balqa` | KP418769 | Morphology and molecular |
| KP418770 | ||||
| KP418771 | ||||
| Bemisia tabaci (MEAM1) | Abelmoschus esculentus (L.) Moench | Amman, AL-Balqa`, Al-Mafraq, Jerash, Jordan Valley, Ma`an, Madaba | KC789925- | Morphology and molecular |
| KC789962 | ||||
| Althaea rosea L. | ||||
| Brassica oleracea var. botrytis L. | ||||
| Brassica oleracea var. capitata L. | ||||
| Brugmansia sp. | ||||
| Capsicum sp. | ||||
| Citrullus lanatus (Thunb.) Matsum. & | ||||
| Nakai | ||||
| Cucumis melo var. flexuosus (L.) | ||||
| Naudin | ||||
| Cucumis sativus L. | ||||
| Cucurbita pepo var. melopepo L. | ||||
| Harz. | ||||
| Cucurbita pepo var. pepo L. | ||||
| Euphorbia pulcherrima Willd. ex | ||||
| Klotzsch | ||||
| Gossypium sp. | ||||
| Helianthus annuus L. | ||||
| Lagenaria siceraria (Molina) Standl. | ||||
| Lantana camara L. | ||||
| Lycopersicon esculentum Mill. | ||||
| Ocimum basilicum L. | ||||
| Ricinus communis L. | ||||
| Solanum melongena L. | ||||
| Solanum tuberosum L. | ||||
| Siphoninus phillyreae | Punica granatum L. | Al-Balqa` | Morphology | |
| Tetraleurodes neemani | Punica granatum L. | Amman, Al-Balqa` | Morphology | |
| Trialeurodes lauri | Arbutus andrachne L. | Jerash | KP418768 | Morphology and molecular |
| Trialeurodes ricini | Ricinus communis L. | Al-Balqa` | KP418777 | Morphology and molecular |
Molecular and phylogenetic analysis
For the purpose of barcoding of the collected whitefly samples, mtCOI gene was amplified and analysed. Around 816 bp fragment of the mtCOI gene was amplified for all the collected samples of whitefly species and B. tabaci species complex (Fig. 1 A). Representative samples of the different whitefly species and B. tabaci were sent for sequencing. Additionally, this PCR product was subjected to CAPS for further analysis of B. tabaci samples.
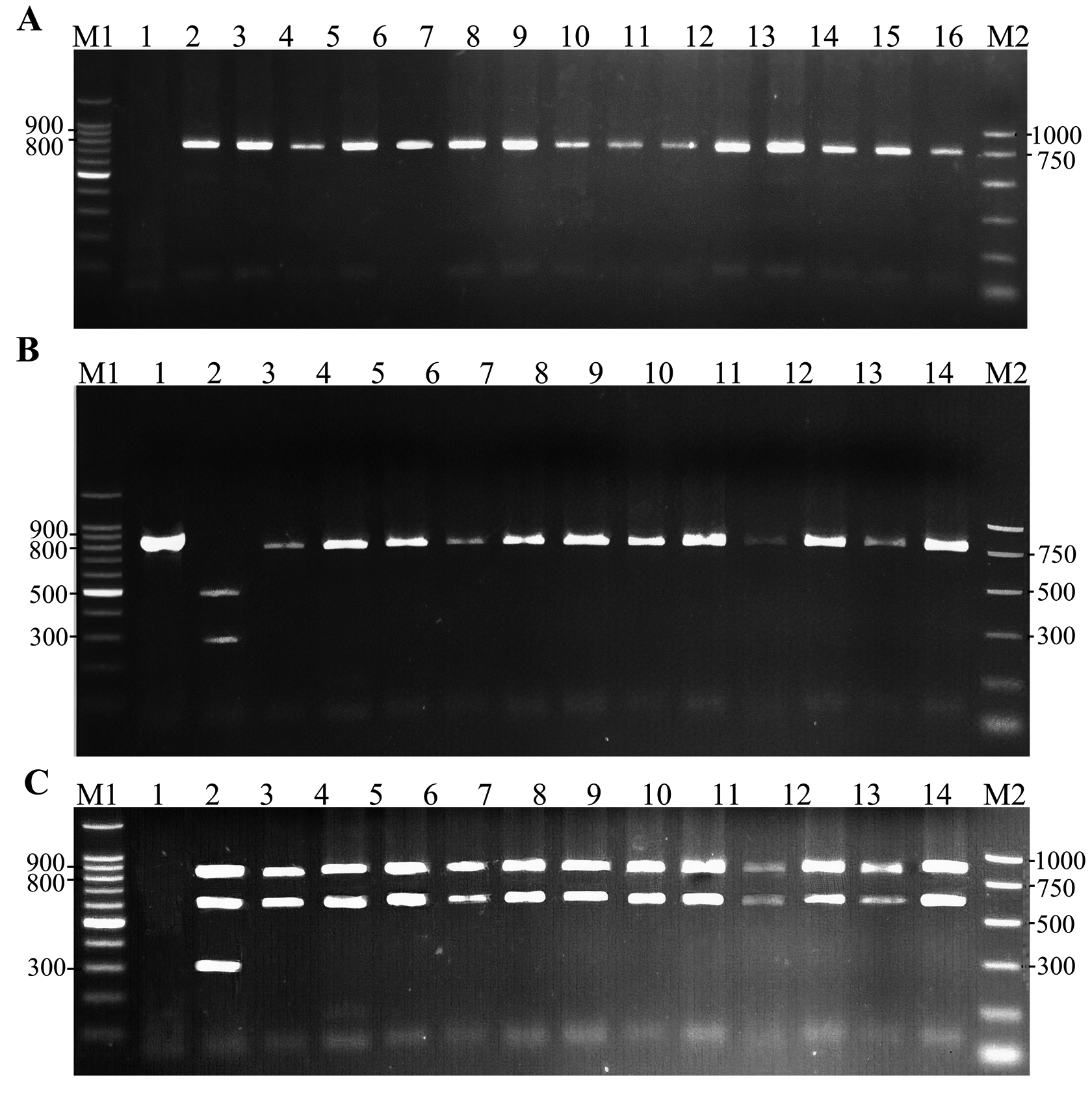
Figure 1 Amplification and analysis of mtCOI. A. Amplification of 816 bp of mtCOI gene for the whitefly species including Bemisia tabaci species complex. Sample 1, negative control; 2, positive control; 3-16, amplified part of mtCOI gene. B. CAPS applied on the amplified mtCOI gene (816 bp) for B. tabaci species complex. Sample 1, undigested PCR product; 2, CAPS pattern of MED reference; 3, CAPS pattern of MEAM1 reference; 4-14, samples of B. tabaci from Jordan. C. The bidirectional PCR analysis. Sample 1, negative control; 2, MED reference; 3, MEAM1 reference; 4-14, samples of B. tabaci from Jordan. Samples were analysed on 1 % agarose gel stained with 0.5 µg/ml of ethidium bromide M1, 100 bp DNA ladder; M2, PCR markers
CAPS and the bidirectional PCR for mtCOI sequence revealed the presence of only MEAMI. In spite of the different hosts, geographical regions, and the time of collection; all the collected B. tabaci samples during the period 2009-2012 showed MEAM1 pattern in CAPS (Fig. 1 B). The same results were confirmed by the bidirectional PCR, the second DNA marker used as it is presented (Fig. 1 C).
After the morphological identification of the whitefly species that were collected from Jordan, the sequences of mtCOI for some of these whiteflies -except B. tabaci- were submitted to GenBank under the accession numbers: KP418768-KP418780. Whereas representative samples of B. tabaci mtCOI sequences were submitted to GenBank under the accession numbers: KC789925-KC789962. All of the B. tabaci samples showed high identity with MEAM1 putative species throughout the world.
To illustrate the relationship within the collected whiteflies samples from Jordan and the other related whiteflies from the world, two phylogenetic trees were constructed. The first tree illustrated the relationship among the differ-ent whitefly species from Jordan including B. tabaci species complex and the other related whitefly species from the world that are available in the GenBank (Fig. 2). The tree confirmed the results obtained from the morphological and molecular identifications. The second phylogenetic tree was constructed for the B. tabaci species complex only (Fig. 3). It showed that all the samples from Jordan are very closely related to the MEAM1 putative species from around the world such as MEAM1 from Japan, Morocco, and Cuba. This as well confirms that all the collected B. tabaci samples from Jordan are MEAM1.
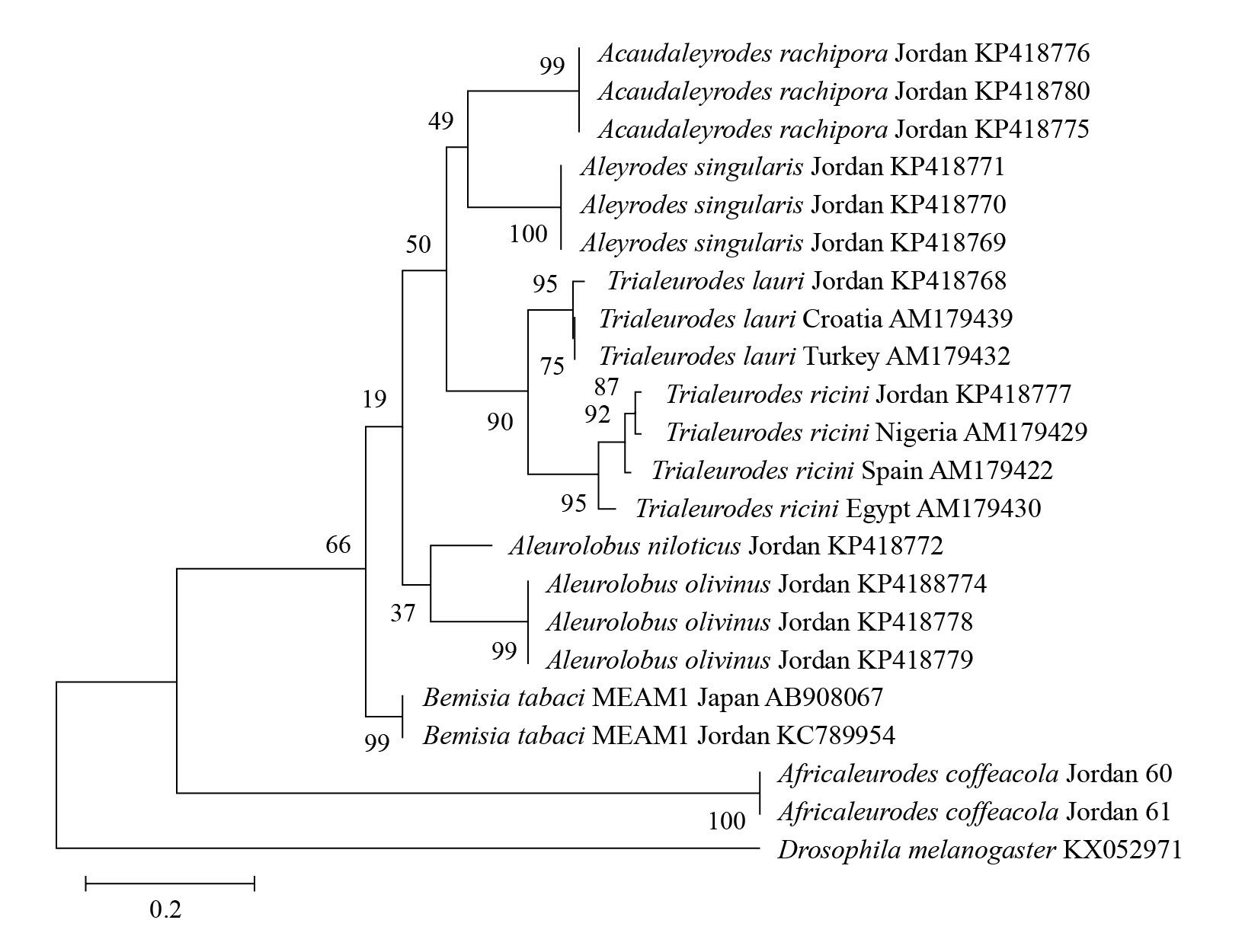
Figure 2 Phylogenetic tree analysis by Maximum Likelihood method based on the Tamura-Nei model for a part of mtCOI gene for different whitefly species collected from Jordan and other parts around the world, Drosophila melanogaster (Meigen,1830) was used as an outgroup taxon. The number at the nodes represents the bootstrapping value and the scale represents the genetic distance. The analyses were conducted using MEGA-X (Kumar et al. 2018).
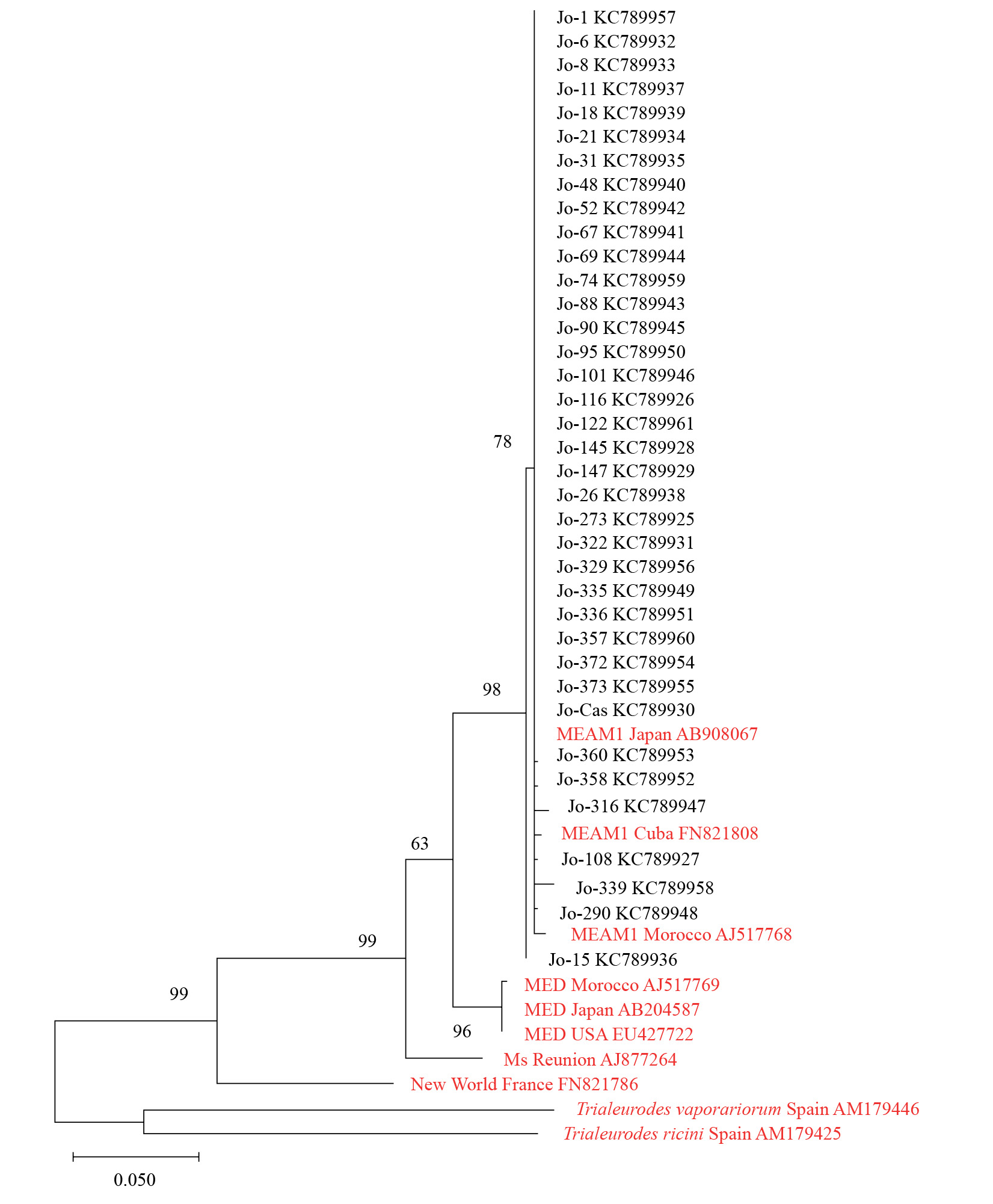
Figure 3 Phylogenetic analysis by Maximum Likelihood method based on the Tamura-Nei model, for a part of mtCOI gene for Bemisia tabaci samples collected from Jordan indicated with (Jo-), the other B. tabaci from the world are colored in red. Trialeurodes vaporariorum and Trialeurodes ricini were used as outgroup taxa. The number at the nodes represents the bootstrapping value and the scale represents the genetic distance. The analyses were conducted using MEGA-X (Kumar et al. 2018).
Discusión
This study aimed to combine the classical morphological identification method of the different whitefly species with the molecular DNA barcoding method for faster and easier identification. Ten different whitefly species were identified mainly based on the morphology. This was due to the poor database of the mtCOI that is available for the whitefly spe-cies other than B. tabaci in GenBank. The obtained sequenc-es were submitted to GenBank, so that, in the future, they would be helpful for the purpose of whitefly identification.
Of the ten species, two species were recorded in Jordan for the first time; these species were T. neemani and Africaleu-rodes coffeacola. T. neemani was also reported in Palestine/Israel (Martin and Mound 2007), China and Iran (Wang et al.2016). Whilist, Africaleurodes coffeacola has been reported in Nigeria (Oyelade and Ayansola 2015) and Congo (Martin and Mound 2007).This study especially focused on studying the genetic di-versity of the invasive whitefly B. tabaci species complex using three techniques based on mtCOI gene sequence (bi-directional PCR, CAPS, and sequencing). In the literatures, MEAM1 and New World putative species have been identi-fied in Jordan using only RAPD technique (Sharaf and Hasan 2003). However, in this study the samples collected during three consecutive years on more than 23 different hosts and from different geographical regions in Jordan confirmed the presence of only MEAM1. It is worth mentioning that the samples were collected from the same places where the New World had been reported earlier as well as the fact that MEAM1 putative species is not new to Jordan as it is known to originate from this area (Broadbent et al. 1989; Cheek and Macdonald 1994; De Barro et al. 2011). MEAM1 pu-tative species are polyphagous, and the females are known for their high fecundity and lower immature mortality (Brown 2007; Costa and Brown 1991; Horowitz et al. 2005; Zhang et al. 2005); this may help this putative species to displace any other species present. In addition to the previous points, the agricultural practices in Jordan could favour the dominance of MEAM1 and the displacement of New World putative species.
Conclusions
Nine species of whiteflies in addition to B. tabaci were identi-fied in Jordan. Identification was primarily based on the mor-phology and supported by the sequences of the mtCOI gene. Two species were reported in Jordan for the first time. Depending on the results, it is important to evaluate the damage caused especially by the newly reported species in Jordan, their hosts and if they could transmit any plant viruses. Also, this study was concerned with updating the status of B. tabaci species complex in Jordan to help in designing effective control methods. The results confirmed the presence of only MEAM1 putative species. As a result, it is important to consider the control methods of this pest, since MEAM1 is an invasive pest, that resists many insecticides and is a vector for many important plant viruses.















