Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Revista Colombiana de Ciencias Pecuarias
versão impressa ISSN 0120-0690versão On-line ISSN 2256-2958
Rev Colom Cienc Pecua v.23 n.4 Medellín out./dez. 2010
Kappa casein genotypes and curd yield in Holstein cows¤
Relación entre los genotipos de la kappa caseína de vacas holstein y el rendimiento en cuajada
Relações entre os genótipos da kappa caseína em vacas holstein a rendimento de requeijão
Gema L Zambrano Burbano1*, Zoot; Yohana M Eraso Cabrera1, Zoot; Carlos E Solarte Portilla1, M.Sc, Dr.Sc; Carol Y Rosero Galindo1, Biol, M.Sc, (c). Dr.Sc.
1Breeding Program, University of Nariño. Research Group on Animal Production and Health. School of Animal Sciences. University of Nariño.
(Received: 18 january, 2010; accepted: 21 september, 2010)
Summary
The objective of this study was to relate kappa casein (CSN3) genotypes with curd yield (RC) and total milk protein (PTP) in Holstein cows located in the high tropics in Narino, Colombia. Twenty seven animals were used to establish the mentioned relationships. The genotype of each animal was determined by PCR - SSCP. Variables were analyzed using a linear model which included the fix effects of genotype, lactation stage, and their interaction. Age of the cow and fat percentage in milk were used as covariates. The results for RC indicate no interaction between genotype and lactation stage. Age was not statistically significant (p>0.05), while fat percent and genotype were significant (p<0.05). The Tukey - Kramer test indicated differences between the BB genotype, compared to homozygous AA and heterozygous AB. The BB genotype resulted in the best performance, requiring the least amount of milk to produce one kg of curd. As for protein content, differences were significant (p<0.05) for the effect of genotype and lactation stage: the homozygous BB had the highest percentage of milk protein during the final (third) stage of lactation.
Key words: lactation stage, milk fat, milk processing, PCR-SSC.
Resumen
El presente estudio tuvo como objetivo determinar las relaciones entre los genotipos para Kappa Caseína (CSN3), el rendimiento industrial en cuajada (RC) y el porcentaje total de proteína (PTP) en vacas Holstein del Trópico Alto de Nariño-Colombia. El genotipo de cada animal fue determinado molecularmente con la técnica PCR - SSCP. Para establecer las relaciones antes indicadas se utilizaron 27 unidades experimentales. Las variables fueron analizadas mediante un modelo lineal en el que se incluyeron los efectos fijos del genotipo, el tercio de lactancia, la interacción entre estos dos factores y como covariables, la edad del animal y el porcentaje de grasa en la leche. Los resultados para RC indicaron que no existe interacción entre los genotipos y el tercio de lactancia. La edad del animal no fue estadísticamente significativa (p>0.05), mientras que la covariable porcentaje de grasa y el genotipo resultaron significativos (p<0.05). La prueba estadística de Tukey - Kramer indicó diferencias entre el genotipo BB, respecto al homocigoto AA y al heterocigoto AB, siendo el primero el de mejor rendimiento, al requerir la menor cantidad de leche para producir un kilogramo de cuajada. En cuanto al porcentaje de proteína, se encontraron diferencias estadísticamente significativas (p<0.05) únicamente por efecto del genotipo y del tercio de lactancia, siendo el homocigoto BB el que presentó mayor porcentaje de proteína en el tercer tercio de lactancia.
Palabras clave: industrialización láctea, PCR-SSCP, porcentaje de grasa, tercio de lactancia.
Resumo
Este estudo teve como objetivo determinar as relações entre os genótipos para Kappa Caseína (CSN3), a rendimento industrial em requeijão (RC) e a percentagem de proteína total (PTP) em vacas Holstein do trópico alto do Nariño Colômbia. O genótipo de cada animal foi determinado molecularmente mediante a técnica PCR-SSCP. Para estabelecer as relações descritas acima, foram utilizadas 27 unidades experimentais e um modelo linear que incluiu os efeitos fixos do genótipo, o terço da lactação, a interação entre estes dois fatores, a idade do animal e o percentual de gordura no leite como covariáveis. Os resultados para (RC) indicaram ñao existe interação entre os genótipos e o terço da lactação, a idade do animal não foi estatisticamente significativa (p>0.05), mas a covariável percentagem de gordura, e o genótipo foram estatisticamente significativos (p<0.05). O teste estatístico de Tukey - Kramer indicou as diferenças entre o genótipo BB em relação a o homozigoto AA, heterozigoto AB, apresentando o primeiro o melhor desempenho porque precisou a menor quantidade de leite para produzir um quilo de requeijão. Em relação ã percentagens de proteína foram encontradas diferenças estatisticamente significativas (p<0.05) pelo efeito do genótipo e do terço de lactação, sendo o homozigoto BB o que a presenteou maior percentagem de proteína, no terceiro terço da lactação.
Palavras chave: industrialização do leite, PCR-SSCP, porcentagem de gordura, terço da lactação.
¤ To cite this paper: Zambrano B GL et. al. Kappa casein genotypes and curd yield in Holstein cows. Rev Colomb Cienc Pecu 2010; 23:422-428
* Corresponding author: E-mail: mitugema@hotmail.com
Introduction
Most dairy farms in Colombia are grouped in four geographical areas termed competitive regions, according to Resolution number 000012 from 2007, issued by the Ministry of Agriculture and Rural Development of this country. The majority of the high-tropics herds, which are mostly specialized dairy farms, are located in Antioquia, Cundinamarca and Nariño provinces, while dual-purpose dairy farms are more abundant in the basins of lowtropics (Agrocadenas, 2006).
One of the most limiting factors affecting competitiveness of specialized dairy farms in Narino is the low quality of the product, in terms of milk composition. According to González et al. (2004), the low protein content in milk negatively affects the efficiency of industrial processes to produce curd and cheese. Therefore, improvement of milk composition in this area is an important matter to take into account when planning nutrition and genetic variables for optimal animal production (Solarte et al., 2009).
The compositional quality of milk depends not only on environmental factors, but also on genetic traits such as the Kappa Casein gene (CSN3), which has been widely studied in order to establish relationships between its polymorphisms with the percent of total milk protein and industrial yield (Naranjo et al., 2007). Research conducted in various regions have reported mixed results regarding the associations between AA, AB and BB genotypes of CSN3 and milk yield (Ng-Kwai-Hang et al., 1984; Mclean et al., 1982; Requena et al., 2007). However, given the complexity of gene expression, it is necessary to compare these results with those obtained under environmental conditions typical of this area in Colombia.
Therefore in this study we established the relationships between CSN3 genotypes and curd yield and percentage of milk protein in Holstein cows that are the predominant breed in the Andean region of Nariño.
Materials and methods
Type of study
The present study was conducted in the municipality of Pasto, in southwestern Colombia, located in north latitude 1°13'22'', west longitude 77°16'22'', at a height of 2690 m, and an average temperature of 12 °C and a relative humidity of 82% (IGAC, 2006).
A total of 348 cows were sampled to determine their CSN3 genotype using PCR-SSCP. Once identified by this criterion, lactating cows were selected and classified according to their lactation stage, i.e., initial (first), mid (second), and final (third).
Methods
Identification of genotypes. Molecular identification of AA and BB homozygous and AB heterozygous genotypes for the CSN3 gene was conducted at the Animal Breeding laboratory of the University of Nariño, following the methodology described by Barroso et al. (1998), and modified by Solarte et al. (2009).
Collection of milk samples. Milk samples were obtained from animals whose output was adjusted to 305 days and adult equivalent (PL) using the factors indicated by Cerón et al. (2003) for Colombian Holstein cattle. The experimental unit was each animal in its corresponding lactating stage. Four liters of milk were obtained from each cow in the morning milking and immediately transported under refrigeration (4 °C) to the processing plant. Upon receipt of the sample, three litters of milk were used for the assessment of curd yield, and one litter for physicochemical, microbiological and compositional analysis.
An EKOMILK (KAM Milkama 98-2A) industrial milk analyzer was used to assess milk composition. Values obtained for acidity and fat using the analyzer were confirmed by manual testing. Acidity was determined by a qualitative change in color after mixing one volume of milk with an alkaline solution of sodium hydroxide, adding phenolphthalein as indicator (0.1 N). Fat content was determined using Gerber's method (1955). The number of colony-forming units (CFU) and degree of environmental contamination of milk were included in the microbiological analysis.
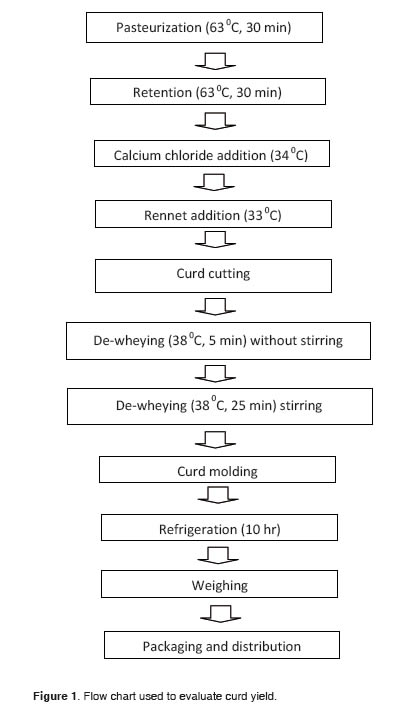
Curd. A protocol was established at the processing plant for the evaluation of curd yield (Figure 1). The equipment and implements used to obtain the curd were a centrifuge (Funke Gerber, Berlin Germany), autoclave (All American, Wisconsin, USA), stainless steel containers, electric stoves, burettes (Schott, Mainz, Germany), pipettes (Schott , Mainz, Germany), thermo-hydrometer (Funke Gerber, Berlin Germany), analytical balance (Mettler Toledo, Mexico DC), thermometers (B & S, Germany), curd knives to cut 1 x 1 cm, industrial molds (500 g), and bags for packaging.
The curd obtained was classified as "peasant type", according to the processing plant. This product is characterized by 60% moisture content. Fat and protein contents were not standardized prior to milk processing.
Evaluation of curd yield. After 10 hours of cooling, the curd yield was calculated taking into account the volume of processed milk and the final weight of the curd, and calculations were based on the following formula:
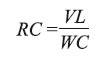
where RC = curd yield, expressed in liters of milk required to produce one kg of curd; VL = volume of milk; and WC = final weight of curd.
Statistical analysis. Analyses were performed using a linear model which included fixed effects of lactation stage (lactation was divided into thirds), genotype, and the interaction effect of lactation stage by genotype. Percentage of fat in milk was included as a covariate. The statistical model is expressed as:
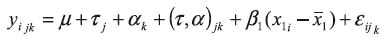
where Yijk = curd yield, associated with the jth genotype, the kth lactation stage, and the interaction of the jth genotype with the kth lactation stage, and taking into account the percentage of fat in milk. μ = Media common to all observations.
τj = Effect of the jth genotype. J= 1, 2, 3.
αk = Effect of the kth lactation stage.
k = 1, 2, 3.
τα = Interaction effect of the jth genotype with the kth lactation stage.
β (x1i – x1) = Lineal effect of the covariate "percent of fat in milk".
εijk = Experimental error associated with the jth genotype in the kth lactation stage and the interaction between the jth genotype with the kth lactation stage.
Each genotype in each lactation stage was regarded as a treatment. Each treatment had three replicates and thus in total 27 experimental units were evaluated. Age was not included in the model as a covariate because it was not statistically significant. The analysis was performed with the GLM procedure of SAS statistical software version 9.20 and Enterprise SAS Guide version 4.2. (2009).
Results
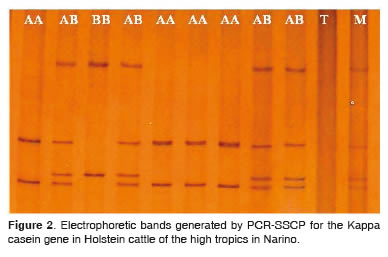
Kappa - Casein (CSN3) genotypes According to Barroso et al. (1998), the PCR SSCP molecular technique identifies more than four allelic variants for the CSN3 gene. In our studied population only two allelic variants were found (A and B), identifying homozygous genotypes AA, BB and heterozygous AB, in accordance with the electrophoretic pattern described by Barroso et al. (1998) (Figure 2). These results are consistent with those reported by other researchers, in which A and B alleles have the highest frequency for the CSN3 gene in dairy breeds (Requena et al., 2007; Chessa et al., 2007; Soria et al., 2003; Eigel et al., 1984).
Descriptive statistics
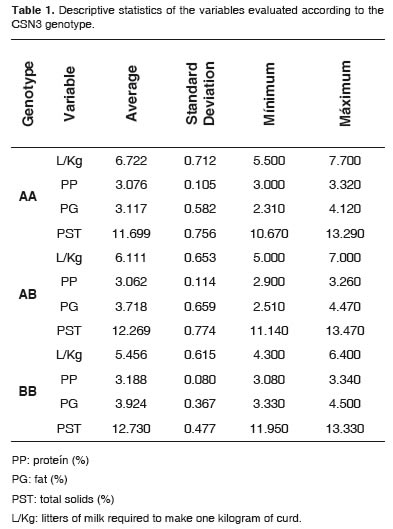
Table 1 presents the percentages of protein (PP), body fat (BF), total solids (PST), and liters required to produce one kg of curd (L / Kg), separated for each genotype.
Analysis of variance for curd yield.
Genotype, lactation stage, and the linear effect of fat percent in milk (included as a covariate) were statistically significant (p <0.05) in the ANOVA. The coefficient of determination was 0.624, indicating that the effects included in the model explain curd yield by 62, 4%.
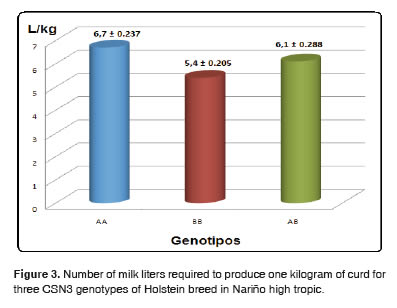
The least square means for curd yield were compared using the Tukey - Kramer test, concluding that the BB genotype had the highest yield compared to genotypes AA and AB, and no significant differences were found between AA and BB, as shown in Figure 3.
Finally, according to the Tukey -Kramer multiple comparison test, the only differences found for protein percentage were between the first and the second third of lactation. This is in agreement with reports by Comeron et al. (2003). However, no differences were observed among the different genotypes.
Discussion
The results of this study are consistent with those reported by Ng-Kwai-Hang et al. (1984) and McLean et al. (1982), who concluded that the BB genotype for CSN3 determines the best milk properties for cheese production, because of the greater firmness in the curd and less time required for the formation of small micelles. In regard to the homozygous AA genotype, these animals had lower casein contents and, as a consequence, a higher proportion of large micelles, which reduces curd yield efficiency.
Furthermore, according to Barroso et al. (1998), there is a positive relation between the CSN3 genotype and the milk protein content, and this protein content influences the clotting time required by rennet, and also the firmness and the cheese yield, showing higher values in milk from cows with genotype BB with respect to the homozygous AA (Schlieben et al., 1991; Van Eenennaam and Medrano, 1991), reaching differences that in some cases amount up to 3% (Aleandri et al., 1990).
The study by Rojas et al. (2009) using Limonero cattle indicated that variant B of the CSN3 could be used to improve the efficiency to transform milk into cheese. To a similar conclusion arrived Naranjo et al. (2007) for the same variant of the gene in Harton del Valle cattle breed. In summary, as stated by Solarte et al. (2009), these studies found that bovine milk from cows with the BB genotype for CSN3 has greater stability to heat and freezing, requires less clotting time, produces a more consistent curd, and increases cheese yield. Those results are in agreement with the present study.
These results confirm the need to reorient the selection process in Nariño high tropics, where CSN3 genotype should be considered as an important criterion to define selection objectives.
This trait should be included along with other factors that are relevant to the region, such as the functional type, longevity and somatic cell count, consistent with current selection trends for the Holstein breed in many countries (Heins et al., 2006).
Acknowledgements
We would like to acknowledge the Colombian Ministry of Agriculture and Rural Development, the Breeding Program at the University of Nariño, and Narino Dairy Cooperative Dairy -Colácteos, for providing the resources necessary for the completion of this study.
References
1. Aleandri RLG, Buttazzoni JC, Schneider A and Davoli R. B. the effects of milk protein polymorphisms on milk components and cheese-production ability. J Dairy Sci 1990; 73:241-255. [ Links ]
2. Barroso A, Dunner S, Cañon J. Detection of Bovine Kappacasein Variants A, B, C y E by Means of Polymerase Chain Reaction-single strand conformation polymorphism (PCRSSCP). Technical note. J Anim Sci 1998; 76: 1535-1538. [ Links ]
3. B.S.I BRITISH STANDARDS INSTITUTTION. Method for determination of fat in milk and milk products. Gerber. 1955. [ Links ]
4. Cerón M, Tonhati H, Costa C, Solarte C, Benavides C. Factores de ajuste para producción de leche en bovinos Holstein colombiano. Colombia Rev Colomb Cienc Pecu 2003; 16: 26-32. [ Links ]
5. Chessa S, Chiatti F, Ceriotti G, Caroli A, Consolandi C, Pagnacco G, Castiglioni B. Development of a Single Nucleotide Polymorphism Genotyping Microarray Platform for the Identification of Bovine Milk Protein Genetic Polymorphisms. J Dairy Sci 2007; 90:451-464. [ Links ]
6. Comerón EA, Aronna MS, Roggero M, Brizi N, Romero LA, Cuatrín A. Sistema de pastoreo líderes-seguidores con vacas lecheras a dos niveles de suplementación. Rev Arg d Prd Anm 2003; 23 Suppl 1:103-104. [ Links ]
7. Eigel WN, Butler JE, Ernstrom CA, Farrell HM, Harwalkar VR, Jenness R, McL Whitney R. Nomenclature of Proteins of Cow›s Milk: Fifth Revision. J Dairy Sci 1984; 67:1599-1631.
8. González MF, Barrales V y Valenzuela AS. Desarrollo de un modelo para evaluar la factibilidad productiva y económica de la progenie resultante del cruzamiento de vacas holstein friesian con toros de las vacas francesa montbeliarde y normando. [Fecha de acceso a la web 2005] URL: http://www.uc.cl/agronomia/e_publicaciones/Documentosderabajo/doc [ Links ]
9. Heins BJ, Hansen LB, Seykora AJ. Production of Pure Holsteins versus crossbreds of Holstein with Normande, Montbeliarde, and Scandinavian Red. J Dairy Sci 2006; 89:2799-2804. [ Links ]
10. IGAC. Instituto Geográfico Agustín Codazzi. Colombia. 2006. URL: www.igac.gov.co/ [ Links ]
11. Mclean DM, Graham ERB and Mckenzie HA. Estimation of casein composition by gel electrophoresis. Int Dairy Congr Moscow 1982; 1:221. [ Links ]
12. Ministerio de Agricultura y Desarrollo Rural Colombia, resolución número 000012 del 2007 "por la cual se establece el sistema de pago de la leche cruda al productor". 12 de enero 2007. [ Links ]
13. Naranjo J, Posso A, Cárdenas H, Muñoz JE. Detección de la variantes alélicas de la kappa caseína en bovinos hartón del valle. Rev Acta Agronóm 2007; 56 :43-47. URL www.revistas.unal.edu.co [ Links ]
14. Ng-Kwai-Hang KF, Hayes JF, Moxley JE, Monardes HG. Association of genetic variants of casein and milk serum proteins in milk fat and protein production by dairy cattle. J Dairy Sci 1984; 67:835-840. [ Links ]
15. Observatorio Agrocadenas. Segundo informe de coyunturas de leche. 2006. Disponible en internet www.agrocadenas.gov.co [ Links ]
16. Requena FD, Agüera EI, Requena F. Genética de la caseína de la leche en el bovino Frisón. Redvet Rev electr Vet 2007; Vol VIII: 1. URL http://www.veterinaria.org/revistas/redvet/ n010107.html. [ Links ]
17. Rojas I, Aranguren MJ, Portillo M, Villasmil OY, Valbuena E, Rincón X, Contreras G, Yañez L. Polimorfismo genético de la kappa-caseína en ganado criollo limonero. Revista científica 2009; 19:645-649. [ Links ]
18. SAS. Institute INC versión 9.20 y Enterprise SAS Guide versión 4.2. 2009. [ Links ]
19. Schlieben S, Erhardt G, Senft B. Genotyping of bovine kappacaseine following DNA sequence amplification and direct sequencing of kappacaseine-E PCR. Anim Genetics 1991; 22:333-342. [ Links ]
20. Solarte C, Rosero C, Eraso Y, Zambrano G, Cárdenas H, Burgos W. Frecuencias alélicas del gen Kappa caseína en la raza Holstein del trópico alto de Nariño-Colombia. Livestock Research for Rural Development 2009; 21(1). URL http://www. lrrd.org/lrrd21/1/sola21003.htm [ Links ]
21. Soria LA, Iglesias GM, Huguet MJ, Mirande SL. A PCR-RFLP Test to detect allelic variants of the bovine kappa-casein gene. Anim Biotechnol 2003; 14:1-5. [ Links ]
22. Van Eenennaan A, Medrano J. Genetic polymorphism in milk protein loci: Milk protein polymorphisms in California dairy cattle. J Dairy Sci 1991; 4:1730-1742. [ Links ]