Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Colombiana de Ciencias Pecuarias
Print version ISSN 0120-0690On-line version ISSN 2256-2958
Rev Colom Cienc Pecua vol.24 no.2 Medellín Apr./June 2011
Artículos originales
HSP90 is a target protein for ubiquitination in Giardia intestinalis¤
HSP90 es una proteina blanco de ubiquitinación en Giardia intestinalis
HSP90 é uma proteína alvo de ubiquitinação em Giardia intestinalis
Jenny J Chaparro-Gutiérrez1,2, MV MS; Moisés Wasserman1*, Quim PhD
1Laboratorio de Investigaciones Básicas en Bioquímica, Facultad de Ciencias,Universidad Nacional de Colombia, Bogot á, Colombia.
2Facultad de Ciencias Agrarias, Escuela de Medicina Veterinaria, Universidad de Antioquia, Medellín, Colombia
(Recibido: 4 febrero, 2011; aceptado: 21 marzo, 2011)
¤ To cite this article: Chaparro-Gutiérrez JJ, Wasserman M. HSP90 is a target protein for ubiquitination in Giardia intestinalis. Rev Colomb Cienc Pecu 2011; 24:107-115
* Corresponding author: Moisés Wasserman. Carrera 45 No 26-85. Unidad Camilo Torres, Bloque 10, Nivel 4, Bogotá D.C., Colombia. E-mail: mwassermannl@ unal.edu.co.
Summary
Previous studies have demonstrated the existence and expression of genes essential to the process of protein ubiquitination in Giardia intestinalis, indicating that the ubiquitin-proteasome system may be involved in the degradation of proteins during its life cycle of the parasite. In this study, purification of ubiquitin was conducted from protein extracts of G. intestinalis trophozoites. Then, an anti-ubiquitin specific antibody was obtained to standardize an assay for the detection and evaluation of ubiquitination patterns. Finally, HSP90 was identified as an ubiquitinated protein in this protozoan. This post-translational modification could have regulatory effects associated with the functionality of the protein or its turnover to regulate key molecular events during the parasite’s life cycle.
Key Words: protozoan, thermic shock protein, ubiquitin.
Resumen
Estudios previos han demostrado la existencia y expresión de genes esenciales para el proceso de ubiquitinación de proteínas en Giardia intestinalis, indicando que el sistema ubiquitina-proteosoma puede estar involucrado en el proceso de degradación de proteínas de este parásito durante su ciclo de vida. En el presente trabajo se realizó la purificación de ubiquitina a partir de extractos proteicos de trofozoítos de G. intestinalis, se produjo un anticuerpo anti-ubiquitina específico que permitió la estandarización de un ensayo para la detección y evaluación de los patrones de ubiquitinación, y se identificó la HSP90 como una proteína ubiquitinada en este protozoario. Esta modificación post-transduccional puede tener efectos regulatorios asociados con la funcionalidad de la proteína o con el recambio para regular eventos moleculares claves durante el ciclo de vida del parásito.
Palabras clave: protozoario, proteína de choque térmico, ubiquitina.
Resumo
Estudos anteriores demonstraram a existência e expressão de genes essenciais para o processo de ubiquitinação de proteínas em Giardia intestinalis, indicando que o sistema ubiquitina-proteassoma podem estar envolvidos na degradação de proteínas do parasita durante seu ciclo de vida. Neste trabalho, foi realizada a purificação da proteína ubiquitina de extratos de trofozoítos de G. intestinalis, foi produzido um antiicorpo anti-ubiquitina específico que permitiu a padronização de um ensaio para a detecção e avaliação de padrões de ubiquitinação e foi identificado a HSP90 como uma proteína ubiquitinada no protozoário. Esta modificação pós-transducional pode ter efeitos regulamentares associados com a funcionalidade das proteínas ou com o a substituição para regular eventos moleculares importantes durante o ciclo de vida do parasita.
Palavras-chave: protozoários, proteínas de choque térmico, ubiquitina.
Introduction
The protozoan parasite Giardia intestinalis (syn. G. lamblia, G. duodenalis) is the principal non-viral causing agent of human diarrheic disease throughout the world (Adam, 2001). In addition to its importance as a pathogen, this eukaryote is considered an important model for the study of basic cellular processes (Morrison et al., 2007). During its life cycle, G. intestinalis presents two differentiation processes, encystation and excystation. Thus, the parasite alternates between an asexually replicating vegetative form (trophozoite) and an infectious form that is able to survive in the external environment (cyst) (Lujan et al., 1997). Both of these processes involve intracellular modifications, transcription of new genes; synthesis, and probably specific degradation of proteins (Adam, 2001).
The ubiquitin/proteasome system (UPS) serves as the most important protein degradation pathway in eukaryotes. Ubiquitin is a small globular protein that is covalently attached to target proteins, most commonly at lysine residues. The covalent attachment of ubiquitin to target proteins is catalysed in a three-step enzymatic cascade involving ubiquitin-activating (E1) and ubiquitinconjugating (E2) enzymes and ubiquitin ligases (E3) (Glickman and Ciechanover 2002). The removal of ubiquitin occurs in one step, catalysed by deubiquitinases (Reyes-Turcu et al., 2009). Historically, the post-translational modification of proteins by ubiquitin has been associated with protein degradation (DeMartino and Gillete 2007). More recently, non-proteolytic functions of ubiquitination have been revealed in many cellular pathways including membrane trafficking, DNA repair and replication and gene transcription (Welchman et al., 2005; Chen and Sun 2009).
Previous studies have demonstrated the existence and expression of essential genes for the ubiquitination process of proteins in G. intestinalis (Gallego et al., 2007; Catic et al., 2007). A ubiquitinated substrate, glucosamine-6-phosphate isomerase has also been identified during the parasite’s encystation process (López et al., 2002). This information suggests that the selective intracellular protein degradation system or ubiquitin-proteasome system could be involved in the protein degradation process in G. intestinalis during its life cycle. With this study was initiated the characterization of the modification caused by the ubiquitin conjugation in G. intestinalis proteins. First, purifying ubiquitin from trophozoites’ protein extracts to produce a specific antibody against conjugates ubiquitin-protein. Then, evaluating the ubiquitination patterns on the trophozoite stage and during the encystation process using an immunoblotting assay and finally, identifying an ubiquitinated protein. This post-transductional modification could have regulatory effects associated with protein’s functionality or turnover to regulate key molecular events driving the parasite life cycle.
Materials and methods
Parasite culture
G. intestinalis trophozoites (WB, Clon C6) were cultured in Diamond TYI-S-33 pH 7.0 medium, supplemented with 10% of bovine serum (Gibco. Invitrogen Corp. NY) and 0.5 mg/mL bovine bile (Sigma, St Louis, MO) at 37 °C in borosilicate tubes (BD Falcon, Biosciences, CA). The tubes were filled with medium in order to get an anaerobic atmosphere and they were kept at an inclination of 45 degrees (Keister, 1983). For the in Vitro encystation, the trophozoites were cultured in a TYI-S-33 pH 7.8 medium supplemented with 10 mg/mL of bovine bile (Kane et al., 1991). 1.000 IU/mL of penicillin (Sigma, St Louis, MO) and 1.0 μg/mL streptomycin (Sigma, St Louis, MO) were also added; the pH was adjusted with a NaOH 5 N solution before the sterilization by filtration with 0.45 μm pore-size nitrocellulose membranes. Samples were collected during different stages of the encystation process: 0 h (before the stimulus), 1 h, 3 h, 6 h, 12 h, 24 h and 48 h after the stimulus. The number of cysts and the efficiency of the encystation were calculated by cell count in a hemocytometer.
Obtention of G. intestinalis protein extract
The extract was obtained from large scale (5 L) trophozoite cultures treated as was previously described. The 3-day culture cells were put on ice for 15 min in order to detach the parasites that were adhered to the culture tube’s surface. Subsequently, they were centrifuged at 900 g for 15 min. In order to separated the culture medium, the cells were washed three times with PBS [137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4 (Dibasic) 14.7 mM KH2PO4 (Monobasic) pH 7.4] cold, sterile and centrifuged at 900 g for 15 min. After the third wash, the cells were resuspended in 1 mL of PBS and the cellular count was made by diluting 1:1.000 in PBS. The parasites were counted two times in a hemocytometer and were stored at –80 °C until their processing. The number of cells was calculated as: N=C*f*104, being N the number of cells in 1 mL; C the average number of cells and f the dilution factor.
10X109 trophozoites of G. intestinalis were collected. This cellular pellet of ~ 20 mL was treated with 80 mL of extraction buffer (30 mM Tris-HCl pH 7.5, 20 mM NaCl, 10% glycerol, 0.5% NP40, 10 mM β-mercaptoethanol, 3 mM MgCl2, 1 mM EGTA). This suspension was maintained in agitation at 4 °C for 1 h, and subsequently was centrifuged at 35.000 g for 15 min. The supernatant was recovered and 500 μL of a protease inhibitor mixture were added (Sigma, St Louis, MO), the final inhibitor concentration was: 2 mM AEBSF (4-(-2-aminoethyl) benzenesulphonyl fluoride, 2.8 μM E64, 3 μM pepstatin A, 8 μM bestatin, 0.6 μM leupeptin, 4 μM aprotinin). The protein quantification of the extract was made by the Bradford method (1976), obtaining a protein concentration of 9.38 g/μL.
Tris-tricine SDS-PAGE gels electrophoresis and western blots
In the discontinuous protein electrophoresis with tricine-SDS-PAGE gels (tricine-sodium dodecyl sulfate polyacrylamide electrophoresis) an anode buffer (20 mM Tris-HCl, pH 8.9) and a cathode buffer (10 mM tris-HCl, 10 mM tricine and 0.1% SDS) were used. In the sample buffer 2X (0.1 M tris-HCl, 24% (v/v) glycerol, 8% (w/v) SDS, 0.2 M DTT, 0.02% (w/v) Coomassie blue G-250), Coomassie blue is used as indicator stain instead of bromophenol blue, because it moves in front of the small peptides. The runs were made at 30 V for 30 min followed by 116 V for approximately 2 h. The proteins that were separated by the previous method were transferred to PVDF (polyvinylidene fluoride) membranes at 25 V overnight in transfer buffer (tris base 25 mM, glycine192 mM, methanol 10% (v/v), pH 8.3).
For the immunodetection using western blot, the protocol was followed as it will be described a) blocking of the membrane for 1 h with TBST-M (Tris-Buffered Saline-Tween-Milk: 20 mM Tris-HCl, pH 7.5; 150mM NaCl; 0.1% tween 20; 5% (w/v, low fat milk) buffer; b) incubation for 1 h with the primary antibody anti-ubiquitin produced in rabbit in TBST-M (1:2.000); c) incubation for 1 h in secondary antirabbit IgG biotinyladed antibody (Invitrogen) in TBST-M (1:3.000); d) incubation for 30 min with streptavidine-phosphatase alkaline (Promega) in TBSM (1:3.000). After each incubation, 3 successive 10 min washes were made with the respective incubation buffer. Detection was done with the chromogenic substrates BCIP (5-bromo-4-chloro-3-indolylphosphate) and NBT (nitroblue tetrazolium) (Promega, Madison, WI) for the alkaline phosphatase in the buffer substrate (tris-HCl 100 mM pH 9.0, NaCl 150 mM, MgCl2 1 mM).
Ubiquitin purification
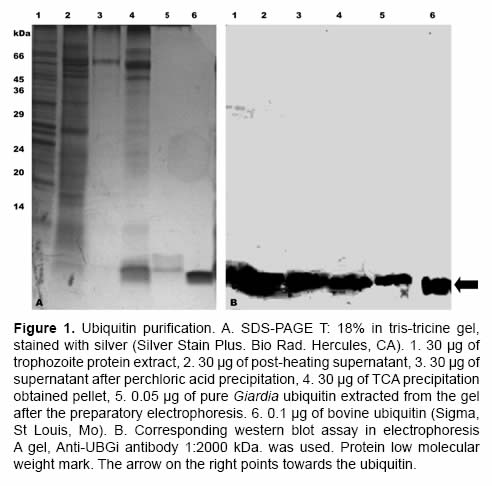
For the purification process was used a modification of the method previously reported by Sparkman and collaborators (1991). Briefly, 100 mL of trophozoite’ protein extract was heated at 85 °C for 20 min in constant agitation; the supernatant was recovered by centrifugation at 30.000 g for 1 h at 4 °C. This solution was precipitated with perchloric acid (final concentration of 5%) in constant agitation for 1 h at 4 °C; the supernatant was recovered as it had been previously done, and this solution was concentrated by precipitation with trichloroacetic acid (final concentration 10%). The pellet was centrifuged at 30.000 g for 5 min at 4 °C; the supernatant was discarded, and the pellet was washed twice with acetone, dried for 30 min at 37 °C and resuspended in 2 mL of deionized sterile water. Lastly, and in order to obtain pure ubiquitin, a preparative extraction from polyacrylamide gels was done following a methodology previously described (Scheer and Ryan, 2001).
Preparative electrophoresis were done in denaturating conditions in tricine-SDS-PAGE gels, running 250 μL samples in buffer 2X (0.1 M Tris-HCl, 24% (v/v) glycerol, 8% (w/v) SDS, 0.2 M DTT, 0.02% (w/v) Coomassie blue G-250). In order to visualize the band and the position of the protein, the extreme of the gel containing the ubiquitin standard and part of the partially purified sample was cut. Then it was stained with Coomassie blue [0.025% (w/v) Coomassie brilliant blue G-250, 10% acetic acid]. According to the electrophoretic patterns the corresponding band was cut; it was disrupted with a 2 cc syringe, and the protein was extracted with the least possible quantity of deionized water maintaining the sample in agitation at 37 °C for 12 h. The process was monitored by vertical electrophoresis in tricine-SDS-PAGE gel and western blot as previously described. The purity of the ubiquitin was verified by tricine-SDS-PAGE. The protein was visualized using silver staining with the Silver Stain Plus (BioRad. Hercules, CA) kit, and was quantified using the Bradford method (1976). This same procedure was done three times.
Anti-ubiquitin antibody production
A specific anti-ubiquitin antibody was produced with the pure ubiquitin following a method that has been previously described (Hershko et al., 1982). Ubiquitin is a small protein (8.5 kDa), it is necessary to cross-link it to a bigger immunogen to generate an immune response; then ubiquitin was coupled to bovine γ-globuline using 3% glutaraldehyde. Bovine γ-globuline (8 mg) and pure G. intestinalis ubiquitin (5 mg) were dissolved in 600 μL of buffer potassium phosphate (0.1 M, pH 7.2). The conjugation of ubiquitin to bovine γ-globuline was made by adding 80 μL of glutaraldehyde to the 3% (v/v) in 20 μL aliquots with 10 minutes intervals in constant agitation. The reaction took place at room temperature for an additional period of 90 min after the last addition of glutaraldehyde. The sample was dialyzed over night against 2 L of PBS pH 7.4 at 4 °C, through 3500 Da cut-off membranes (Spectrum Laboratories, Inc USA). G. intestinalis ubiquitin was denaturated in its final purification stage. The antigen’s denaturation is essential for obtaining an antiubiquitin antibody capable of recognizing ubiquitinprotein conjugates (Hershko et al., 1982).
The polyclonal antibody was generated in a two month old New Zealand rabbit. The inoculation chart proposed by Hershko y collaborators was followed (1982). Briefly, the sample of pure ubiquitin was mixed with an equal volume of Freund’s complete adjuvant (Sigma, St Louis, MO) for the first inoculation and with Freund’s incomplete adjuvant (Sigma, St Louis, MO) (next inoculations- 4 total). The samples containing 0.1 mg of ubiquitin were injected every 2 weeks into rabbits. Antibody titers were determined by western blot assays as described previously. The antibody obtain against G. intestinalis ubiquitin was called Anti-UbGi.
Western blot assays to evaluate ubiquitination patterns
Adequate conditions for a western blot immunodetection assay were standardized with the antiserum (Anti-UbGi). This allowed the analysis of the presence of free ubiquitin and the evaluation of the ubiquitination patterns in trophozoites and during the encystation process of G. intestinalis. For these experiments, protein extracts was prepared as previously described Alvarado and Wasserman (2010) and whole parasites (50.000 cells/μL) were also used, the adequate conditions for the western blot immunodetection assay were also standardized. Some of these conditions are the optimal title of the produced polyclonal antibodies, and the adequate concentration of protein extract or parasite quantity for the specific detection of free ubiquitin or ubiquitinated proteins.
The optimal dilution of the antibodies was established with the detection limit that was evident with pure ubiquitin and the parasite’s protein extracts. For the in vitro encystation, the trophozoites were cultured and collected as it was previously described. The experiments were done by triplicate and with at least two different samples in each experiment. The methodology for electrophoretic and immunodetection assays was the same one that was previously described.
Identification of a ubiquitinated target protein
24 h-trophozoite cultures were treated overnight with 50 μM of the proteasome inhibitor carbobenzoxylleucinylleucinylleucinal-H (MG132) (Calbiochem, La Jolla, CA) in DMSO to enrich the ubiquitin-protein conjugates (Vasilescu et al., 2005). Subsequently, the parasites were harvested as previously described containing a cocktail of protease inhibitors (Sigma, St Louis, MO). The suspension was adjusted to 50.000 cells/μL in sample buffer 1X (15 mM Tris-HCl, pH 6.8, 0.05% (w/v) SDS, 0.002%(w/v) bromophenol % (v/v) glycerol, 89 mM β-mercaptoethanol), heated at 85 °C for 5 min and was evaluated by SDS-PAGE (gels of 8%T, 2.6%C, to resolve the region with high molecular weight proteins) and western blot was done as described.
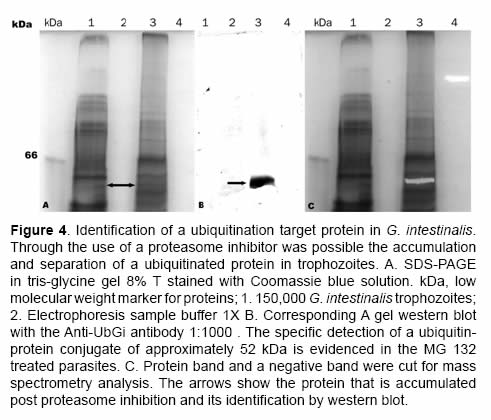
Using the ubiquitination patterns, the identification of the proteins recognized by the antiubiquitin specific antibody was possible. An evident band between 45 kDa and 66 kDa on the parasites that had been treated with the proteasome inhibitor was cut; it was clearly identified by the western blot. The band that was cut had been stained with Coomassie blue and was transferred to low retention Eppendorf tubes. The protein sample was distained by incubation for 2h in 200 μL of a solution of 50% acetonitrile and 50 mM ammonium bicarbonate.
Then, the gel was dehydrated by treatment with acetonitrile 100% for 5 min. The solvent was removed and the drying process at room temperature was allowed. Subsequently, the gel was incubated in a washing solution (50 mM ammonium bicarbonate) for 10 min and dehydrated once more. After this process the gel was treated with 10 mM DTT for 30 min; then the reductor agent was removed and the gel was washed and incubated with 100 mM of iodocetamide for 30 min. The washing procedure was repeated and the gel was left in digestion for 14 h at 37 °C with 5 μL (20 ng/μL) of Trypsin Gold (Promega) in 50 mM of ammonium bicarbonate. After the enzymatic digestion the peptides were eluded from the gel through incubation for 10 min with 30 μL ammonium bicarbonate 50 mM. The first elusion was recovered and the gel was treated two times with 30 μL of an acetonitrile 50% and formic acid 5% mixture. These two elusions were mixed with the first one and concentrated. Finally, the sample was analyzed using the mass spectrometry technique MS+MS/MS. The list of the generated masses was analyzed with the MASCOT software (www.matrixscience.com).
Results
Ubiquitin purification
After preparatory electrophoresis SDS-PAGE with tris-tricine buffer, up to 1 mg of pure ubiquitin was obtained from a culture of 10X109 trophozoites. In figures 1A and 1B the ubiquitin purification process is observed. During the entire process a band with molecular weight of ~ 8.5 kDa was evidenced. It migrated at the same height than bovine commercial ubiquitin (Figure 1, lane 6), the identity of the ubiquitin was proved by a western blot assay (Figure 1B), using an anti-ubiquitin polyclonal antibody.
Ubiquitination patterns
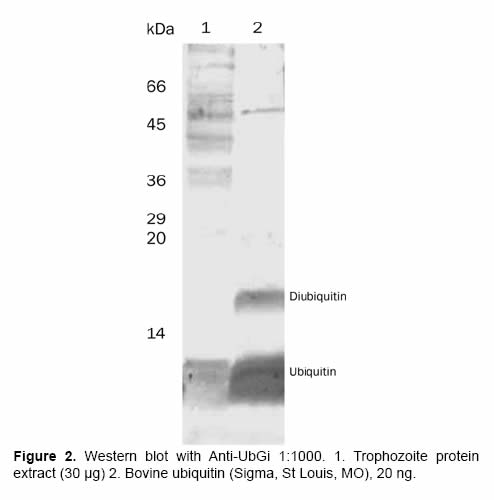
The serum obtained during the rabbit immunization, recognized pure G. intestinalis ubiquitin and commercial bovine ubiquitin (Sigma, St Louis, MO) using western blot assays with 100 ng ubiquitin (Figure 2). These proteins have an identity of 86.84% using protein-protein blast (http://ca.expasy.org). This recognition started 30 days after the first inoculation and the detection increased in time using higher serum dilutions. This evidenced the increase in the antibody titer and in its specificity (data not shown). The pre-immune serum did not detect ubiquitin in any of the studied dilutions. Some variables where determined, one of them was the optimal dilution of the polyclonal antibody when the pure antigen (1:1.000) was used. The protein extract concentration or the right number of parasites for the specific detection of ubiquitin and ubiquitinated proteins was also determined: 100.000 trophozoites and 5 μg of protein extract were enough to detect the ubiquitin and ubiquitin-protein conjugates in G. intestinalis.
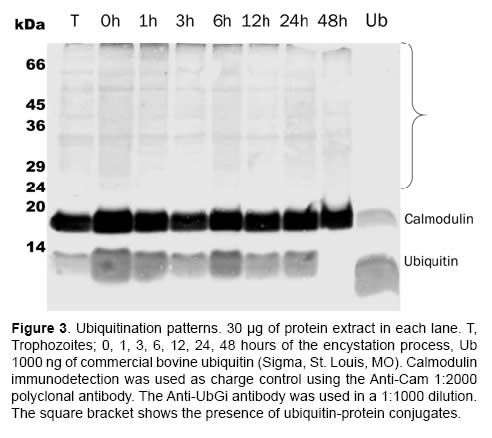
When the presence of ubiquitin-protein conjugates was being evaluated, the anti-ubiquitin policlonal antibody (Anti-UBGi) made it possible to identify the ubiquitination patterns in the high molecular weight region during the trophozoite stage (Figure 3, lane T) and during the encystation process (Figure 3, from 0 to 48 h, see parenthesis). The expression of free ubiquitin was constant until the 24 first hours of the encystation process of G. intestinalis; at 48 h there was an evident decrease of this type of ubiquitin in the soluble cytoplasm fraction (Figure 3, lane 48 h).
Identification of HSP90 as a ubiquitinated protein
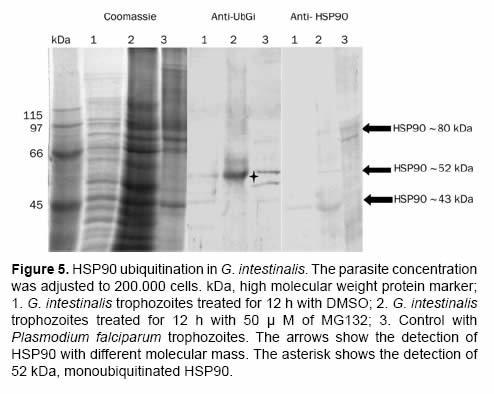
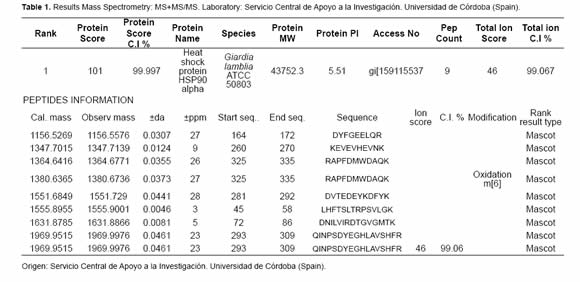
After the proteasome inhibition with MG132 the accumulation of several proteins with molecular masses higher than 50 kDa was evident. A protein between the 45 kDa and the 66 kDa marks (Figure 4 and Figure 5) with low parasite concentration could be observed. This band was cut and processed for its identification through mass spectrometry MS+MS/MS technique and it was identified as “heat shock protein HSP90-alpha (Giardia lamblia ATCC 50803)” with a molecular mass of 43.752 kDa (Figure 6). To corroborate the identity of this protein and validate this result, western blot assays were done using a monoclonal antibody Anti-HSP90 (Figure 5, Anti-HSP90). One of the bands recognized by the monoclonal antibody anti-HSP90 was also clearly marked with the antiubiquitin antibody both in the control trophozoites and the inhibited trophozoites (Figure 5, Anti-UbGi, lanes 1 and 2, respectively) and it corresponded to a ~52 kDa protein. This result indicates that the protein identified by mass spectrometry is the monoubiquitinated (8.5 kDa) HSP90-alpha (43.752 kDa), which is located in exactly 52 kDa (Figure 5, Anti-UbGi, lane 2 and Anti-HSP90, lane 2). With the monoclonal antibody anti-HSP90 a bigger band corresponding to the whole protein can be identified (Figure 5, Anti-HSP90, lane 2).
Discussion
G. intestinalis ubiquitin was purified through a three step method that uses this protein’s thermostability and solubility in the perchloric acid 5% solution. The pure protein made it possible to obtain a specific polyclonal antibody for G. intestinalisubiquitin (Anti-UbGi) which recognized free ubiquitin and ubiquitin-protein conjugates both in soluble protein extracts and in whole parasites. The detection of ubiquitin by the pre-immune serum could not be seen in any of the studied dilutions, confirming that the detection was because of the immunization process.
The decrease in the ubiquitin levels at 48 h after the encystation stimulus in cytoplasm soluble protein extracts suggests that a ubiquitin conjugation to proteins or a relocalization of this protein from the parasite’s cytoplasm to the cyst’s walls could be happening. Stefanic and collaborators (2006) reported the association of the 26S proteasome with the encystation specific vesicles and demonstrated the relocalization of this protease towards the parasite’s periphery during the early stages of G. intestinalis’ encystation. It is possible to suggest that the ubiquitination machinery could go through the same relocalization or that the ubiquitin is linked to proteins related with the periphery of cyst.
The protein identified as ubiquitinated is a cytoplasm protein involved in several cellular processes: the protein degradation in cells exposed to stress, in a key regulatory protein conformational development, and in the growth and development regulation of various protozoan (Pallavi et al., 2010). HSP90 is strongly regulated through several control mechanisms that regulating its activity. Among them are: conformational changes; cofactor presence and post-transductional modifications such as acetylation (Scroggins et al., 2007), S-nitrosylation (Martínez-Ruiz et al., 2005), and phosphorylation (Mollapour et al., 2010).
Even though in various eukaryote cell systems HSP90 has been identified as a ubiquitinated protein (Kirkpatrick et al., 2005; Vasilescu et al., 2005; Morales and Perdew 2007), the physiological role of this modification is not yet known. In our experimental model the detection of HSP90 ubiquitination enhanced in the proteasome inhibited parasites, suggest that its ubiquitination level is too low (Figure 5, Anti-UbGi); an evident level can be reached only after the accumulation of ubiquitinprotein conjugates has been induced (Figure 5, Anti-UbGi, lane 2). It is well documented, in other eukaryote systems, that the cell treatment with proteasome inhibitors causes the induction of a heat shock response which generates the expression of the heat shock proteins (HSP), including HSP90 (Bush et al., 1997; Lee and Goldberg 1998; Mitsiades et al., 2002).
It is important to consider that this type of proteins can be involved in the differentiation process of G. intestinalis. Kim and collaborators (2009) found a peak of messenger RNA for HSP90 at 6 h after the induction of the encystation process. This suggests the expression of these proteins in a differential way during G. intestinalis’ life cycle. On the other hand, it has recently been proved that HSP90 protein in G. intestinalis goes through a post-transcriptional modification to produce the entire ~ 80 kDa protein (Nageshan et al., 2011), making it possible the production of the two fragments of the protein. With this research we have been able to demonstrate that the alpha fraction of G. intestinalis HSP90 is monoubiquitinated. This post-transductional modification could have regulatory effects associated with this protein’s functionality or turnover.
Acknowledgements
The authors want to express their gratitude to the MSc bacteriologist Eva Amanda Gallego for her collaboration with the sample processing through the mass spectrometry technique. This research Project was financed by the Investigation board from Universidad Nacional de Colombia (Projects No 8003151 and 8003083), Bogotá.
References
1. Adam RD. Biology of Giardia lamblia. Clin Microbiol Rev 2001; 14:447-475.
[ Links ]2. Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem 1997; 272:9086-9092.
[ Links ]3. Catic A, Sun ZYJ, Ratner DM, Misaghi S, Spooner E, Samuelson J, Wagner G,Ploegh HL. Sequence and structure evolved separately in a ribosomal ubiquitin variant. EMBO J 2007; 26:3474-3483.
[ Links ]4. Chen ZJ, Sun LJ. Nonproteolytic Functions of Ubiquitin in Cell Signaling. Mol Cel Rev 2009; 33:275-286.
[ Links ]5. Darryl Pappin and David Perkins. Cancer Research Technology, Labvantage Solutions and Swiss-prot. Accession date: November 24 of 2008. URL: http://www.matrixscience.com/ home.htm
[ Links ]6. DeMartino GN, Gillette T. Proteasomes: machines for all reasons. Cell 2007; 129: 659-662. DOI:10.1016/j. cell.2007.05.007.
[ Links ]7. Gallego E, Alvarado M, Wasserman M. Identification and expression of the protein ubiquitination system in Giardia intestinalis. Parasitol Res 2007; 1:1-7.
[ Links ]8. Glickman MH, Ciechanover A .The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 2002; 82:373-428.
[ Links ]9. Hershko A, Eytan E, Ciechanover A. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. J Biol Chem 1982; 257:13964-13970.
[ Links ]10. Kane AV, Ward HD, Keusch GT, Pereira ME. In vitro encystation of Giardia lamblia: large-scale production of in vitro cysts and strain and clone differences in encystation efficiency. J Parasitol 1991; 77:974-981.
[ Links ]11. Keister DB. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R SocTrop Med Hyg 1983; 77:487-488.
[ Links ]12. Kirkpatrick DS, Weldon SF, Tsaprailis G, Liebler DC, GandolfiAJ. Proteomic identification of ubiquitinated proteins from human cells expressing His-tagged ubiquitin. Proteomics 2005; 5:2104-2111.
[ Links ]13. Lee DH, Goldberg AL. Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol Cell Biol 1998; 18:30-38.
[ Links ]14. López AB, Hossain MT, van Keulen H. Giardia intestinalis glucosamine 6-phosphate isomerase: the key enzyme to encystment appears to be controlled by ubiquitin attachment. J Eukaryot Microbiol 2002; 49:134-136.
[ Links ]15. Luján HD, Mowatt MR, Nash TE. Mechanisms of Giardia lamblia differentiation into cysts. Microbiol Mol Biol Rev 1997; 61:294-304.
[ Links ]16. Martínez-Ruiz A, Villanueva L, González de Orduña C, LópezFerrer D, Higueras MA, Tarín C, Rodríguez-Crespo I, Vázquez J, Lamas S. S-nitrosylation of hsp90 promotes the inhibition of its ATPase and endothelial nitric oxid synthase regulatory activities. Proc Natl Acad Sci U S A 2005; 102:8525-8530.
[ Links ]17. Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D,y Fanourakis G.. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci U S A 2002; 99:14374-14379.
[ Links ]18. Mollapour M, Tsutsumi S, Neckers L. Hsp90 phosphorylation, Wee1, and the cell cycle. Cell Cycle 2010; 8:9:2310-2316.
[ Links ]19. Morales GL y Perdew GH. Carboxyl terminus of hsc70interacting protein (CHIP) can remodel mature aryl hydrocarbon receptor (AhR) complexes and mediate ubiquitination of both the AhR and the 90 kDa heat-shock protein (hsp90) in vitro. Biochemistry 2007; 16:610-621.
[ Links ]20. Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ, Davids BJ, Dawson SC, Elmendorf HG, Hehl AB, Holder ME, Huse SM, Kim UU, Lasek-Nesselquist E, Manning G, Nigam A, Nixon JEJ, Palm D, Passamaneck NE, Prabhu A, Reich CI, Reiner DS, Samuelson J, Svärd SG, Sogin ML. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science 2007; 317:1921-1926.
[ Links ]21. Nageshan RK, Roy N, Hehl AB, Tatu U. Post-transcriptional repair of a split heat shock protein 90 gene by mRNA trans-splicing. J Biol Chem in press 2011; http://www.jbc.org/cgi/doi/10.1074/jbc.C110.208389
[ Links ]22. Pallavi R, Roy N, Nageshan RK, Talukdar P, Pavithra SR, Reddy R, Venketesh S, Goel RK, Gupta AK, Singh RK, Yadav SC, Tatu U. Heat shock protein 90 as a drug target against protozoan infections: biochemical characterization of HSP90 from Plasmodium falciparum and Trypanosoma evansi and evaluation of its inhibitor as a candidate drug. J Biol Chem 2010; 285:37964-37975.
[ Links ]23. Scheer JM, Ryan CA. A method for the quantitative recovery of proteins from polyacrylamide gels. Analyt Biochem 2001; 298:130-132.
[ Links ]24. Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, Rosen N, Neckers L. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol Cell. 2007; 25:151-159.
[ Links ]25. Sparkman D, Hill C, White S. Rapid one-step extraction procedure for the isolation of ubiquitin from human erythrocytes for antibody production. Prep Biochem 1991; 21:93-104.
[ Links ]26. Stefanic S, Palm D, Svärd SG, Hehl AB. Organelle proteomics reveals cargo maturation mechanisms associated with Golgi-like encystation vesicles in the early-diverged protozoan Giardia lamblia. J Biol Chem 2006; 281:7595-7604.
[ Links ]27. Swiss Institute of Bioinformatics. Accession date: October 15 of 2008. URL: http://expasy.org/tools/blast/.
[ Links ]28. Vasilescu J, Smith JC, Ethier M, Figeys D. Proteomic analysis of ubiquitinated proteins from human MCF-7 breast cancer cells by immunoaffinity purification and mass spectrometry. J Proteome Res 2005; 4:2192-2200.
[ Links ]29. Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitinlike proteins as multifunctional signals. Nat Rev Mol Cell Biol 2005; 6:599-609.
[ Links ]













