Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Colombiana de Ciencias Pecuarias
Print version ISSN 0120-0690On-line version ISSN 2256-2958
Rev Colom Cienc Pecua vol.25 no.1 Medellín Jan./Mar. 2012
Artículos originales
Assessing the genetic diversity and ancestry of Hartón delValle cattle using mitochondrial DNA¤
Diversidad genética y ancestría del ganado Hartón del Valle mediante ADN mitocondrial
Estimação da ancestralidade e da diversidade genética no gado Hartón del Valle utilizando DNAmitocondrial
Luz A Álvarez1*, Zoot, PhD; Víctor J Vera2, DMV, PhD; Heiber Cárdenas3, MSc Biol; Guillermo Barreto3Biol, PhD; Jaime E Muñoz1, Agr Eng, PhD
1 Professor, Faculty of Agricultural Sciences, National University of Colombia, AA 237, Palmira, Colombia.
2 Professor, Faculty of Veterinary Medicine, National University of Colombia, Bogotá, Colombia.
3 Department of Biology, Faculty of Sciences,University of Valle, AA 25360, Cali, Colombia.
(Recibido:18 junio,2010; aceptado: 22 agosto, 2011)
Summary
The Hartón del Valle(HV)cattleisa Criollo breed of Spanish descent that has adapted to the conditionsof the Cauca valley (Colombia). This breed is currently listed as “vulnerable” because its population has dramatically declined in the last two years. Prompted by the uncertain future of this breed and its importance as a potential resource of valuable genes. Objective: this study addressed the diversity, genetic structure and ancestry of HV cattle using mitochondrial DNA (mtDNA). Methods: a 350-bp fragment was amplified and sequenced from 72 HV animals in 9 separate farms and from animals of Brahman and Holstein breeds. To analyze the breed's ancestry, the sequences were compared with 560 sequences available in the GenBank, representing 50 Bos taurus and Bos indicus breeds. Results: in accordance with the Spanish origin of the HV breed, there was a high representation of European mtDNA (91.7%) and a low representation of African (5.5%) and Middle Eastern mtDNA (2.7%). The average haplotype diversity was 0.65 ± 0.05. The farm with the oldest ancestry was the only population in which three mitochondrial lineages were observed; unfortunately, it was recently depopulated. Proximity was observed between HV and two Columbian breeds, the Romosinuano and Costeño con Cuernos. A comparative analysis with the sequences deposited in the GenBank from numerous breeds revealed the presence of 37 haplotypes, seven of which were unique to HV. Conclusions: the following Iberian breeds were found to be most closely related to HV: Tudanca, Rubia Gallega, Negra Serrana, Murciana, Pajuna, Avileña, Garvonesa and Mertolenga. Phylogenetic analysis confirmed the Iberian ancestry and some African influence on this Latin American Criollo breed.
Key words: creolecattle, haplotypes, lineages, phylogeny.
Resumen
El Hartón del Valle (HV),es una raza adaptada a las condiciones del Valle del Cauca(Colombia) que está catalogada como “vulnerable” y cuya población ha descendido drásticamente en los últimos dos años. Objetivo: debido al futuro incierto y a la importancia como recurso potencial en la seguridad alimentaria regional se abordó el estudio de la diversidad, la estructura genética y la ancestría del HV, mediante ADN mitocondrial (ADNmt). Métodos: se amplificó y secuenció un fragmento de 350 pb en 72 animales HV, provenientes de nueve poblaciones y de controles de las razas Brahman y Holstein. Para analizar la ancestría las secuencias fueron comparadas con 560 secuencias de 50 razas Bos taurus y Bos indicus, depositadas en el GenBank. Resultados: de acuerdo con su origen español, se encontró una marcada influencia del ADNmt europeo (91.7%) y baja participación de taurinos de origen africano (5.5%) y del cercano Oriente (2.7%). La diversidad haplotípica promedio fue 0.65 ± 0.05. El hato más antiguo fue el único que mostró los tres linajes mitocondriales, sin embargo, este ha sido liquidado recientemente. Se observó proximidad entre el HV con las razas colombianas Romosinuano y Costeño con Cuernos. La comparación con las secuencias depositadas en el GenBank con diferentes razas reveló la presencia de 37 haplotipos, de los cuales siete fueron únicos en el HV. Conclusiones: las razas ibéricas más cercanas al HV fueron: Tudanca, Rubia Gallega, Negra Serrana, Murciana, Pajuna, Avileña, Garvonesa y Mertolenga. El árbol filogenético confirmó la ancestralidad ibérica y la influencia africana en las razas criollas de América Latina.
Palabras clave: filogenia, ganadocriollo, haplotipos, linajes.
Resumo
O Hartón del Valle (HV) é uma raça adaptada às condições da região pertencente ao departamento do Valle del Cauca (Colombia). A raça está catalogada como vulnerável já que sua população tem sido reduzida drasticamente nos últimos dois anos. Objetivo: devido a incerteza no futuro da raça e a sua importancia como recurso potencial na segurança alimentaria regional, abordou-se o estudo da diversidade, a estrutura genética e a ancestralidade do gado HV, utilizando DNA mitocondrial (mtDNA). Métodos: para isto, se amplificou e sequenciou um fragmento de 350 pares de bases em 72 animais provenientes de nove populações e também de controles das raças Brahman e Holandesa. Para analizar a ancestralidade das sequências, estas foram comparadas com outras 560 de 50 raças Bos taurus y Bos indicus depositadas no genebank. Resultados: de acordo com sua origem espanhola, foi encontrada uma marcada influência do DNA mitocondrial europeu (91.7%), e baixa participação de taurinos de origem africana (5.5%) e do Oriente Próximo (2.7%). A média da diversidade haplotípica foi de 0.65 ± 0.05. A população de gado HV mais antiga foi a única que apresentou as três linhagens mitocondriais encontradas; embora, esta população foi terminada recentemente. Observou-se também proximidade do gado HV com as raças colombianas da região Caribe, Romosinuano e Costeño con Cuernos. A comparação com as sequências depositadas no genebank com diferentes raças revelou a presença de 37 haplotipos, dos quais sete foram únicos no HV. Conclusões: as raças ibéricas mais aproximadas do HV foram: Tudanca, Rubia Gallega, Negra Serrana, Murciana, Pajuna, Avileña Garvonesa e Mertolenga. A árvore filogenética confirmou sua ancestralidade ibérica e a influência africana nas raças crioulas da América Latina.
Palavras chave: filogenia, gadonativo, haplótipos, linhagens.
¤ To cite this article: Álvarez LA, Vera VJ, Cárdena H, Barreto G, Muñoz JE. Assessing the genetic diversity and ancestry of Hartón del Valle cattle using mitochondrial DNA Rev Colomb Cienc Pecu 2012; 25:14-26.
* Corresponding author: Luz Ángela Álvarez. Department of Animal Science. Faculty of Agricultural Sciences, National University of Colombia at Palmira. AA237, Palmira, Colombia. E-mail: laalvarezf@unal.edu.co.
Introduction
The origin of the Hartón del Valle (HV) cattle breed is similar to that of other Colombian Creole breeds. Iberian cattle brought by the Spanish during the colonial era underwent a long process of natural selection to adapt to local conditions, which is evidenced by the high reproductive rates, tolerance to parasite diseases and increased survival of these breeds (Casas and Valderrama, 1998).
In the last census of Creole cattle, the population consisted of 5,120 purebred animals (Martínez, 1999), and ten years later, estimates indicate that the population was reduced to 3,500 animals. Additionally, three of the four farms that provided breeding stock have closed down in recent years. The decrease in the number of Creole cattle has resulted from the conversion of cattle farms to sugar cane farms, the interbreeding of cattle with foreign breeds and the migration of cattle to other provinces. In particular, the HV breed has never been included in the protection programs for Creole cattle, which were established by the Colombian government in the 1930s.
The analysis of mitochondrial DNA (mtDNA) sequences has been used extensively to study the origins of and the relationships among cattle breeds (Bruford et al., 2003). Although the T3 haplogroup is predominant among American Creole cattle breeds, haplotypes of African origin have also been detected (Cymbron et al., 1999; Miretti et al., 2002; Beja-Pereira et al., 2006; Egito, 2007; Magee et al., 2002; Mirol et al.,2003; Lirón et al.,2006; Ginja etal.,2009).
By studying the mtDNA control region in 110 samplesfromsevenColombianCreolecattle breeds, Carvajal-Carmona et al. (2003) found that 65% of the mitochondrial samples corresponded to European ancestry (T3), 26% to African ancestry (T1) and 9% to the T2 lineage. Using 21 samples from the HV breed, the percentages observed for the T1, T2 and T3 lineages were 38%, 29% and 33%, respectively (Carvajal-Carmona et al., 2003).
The present study was motivated by concerns about the vulnerability and uncertain future of the HV breed, as well as the importance of the breed as a potential resource for regional food security. The aims of the study were to analyze the diversity, genetic structure and ancestry of the HV breed and its relationships with Bos taurus and Bos indicus breeds using mitochondrial DNA analysis.
Materials and methods
Sampling, amplification and sequencing of mtDNA
The mtDNA was obtained from blood samples, which were processed using the desalting method (Miller et al., 1988). Because mtDNA haplotype diversity is low (≤ 0.03), 100 random samples were taken from unrelated animals in addition to 500 HV samples from the DNA bank at the National University of Colombia at Palmira. After analyzing the samples, however, only 72 sequences met the quality standards. These samples came from nine farms located in the Valle del Cauca: El Capricho (EC), Casa Rincón (CR), the National University of Colombia (UN), Gran Capricho (GC), Jamaica (JA), La Ondina (LO), Procampo (PR), San Rafael (SR) and Zanjón Hondo (ZH). The oldest herds and varied production systems (milk, meat and dual- purpose) were represented. As a reference, samples from unrelated Brahman (B; N = 6) and Holstein (H; N = 9) cattle were randomly selected from the DNA bank among 250 samples of these two breeds.
A 350-bp fragment located in the most highly variable region of the D-loop was amplified using the primers AN4 (5'-GGTAATGTACATAACATTAATG-3') and AN3(5'-CGAGATGTCTTATTTAAGAGG-3'),as described by Cymbron et al. (1999). This sequence was selected to analyze the relationships between cattle breeds because it has been widely used in different studies of Iberian breeds and American Creole breeds (Bradley et al., 1996; Beja-Pereira et al., 2006). The amplification reactions were carried out using a 50 µl reaction volume with 50 ng of mtDNA, 0.6 units of Taq polymerase, 10X PCR buffer, 3.5 mM MgCl2, 1.25 mM dNTPs and 20 μM forward and reverse primers. The PCR conditions were as follows: an initial denaturation for 3 minutes; 35 cycles of 94 °C for 20 seconds, 55 °C for 40 seconds and 72 °C for 40 seconds; and a final extension at 72 °C for 4 minutes. During the annealing step of the second amplification cycle, the reaction was paused at 55 °C for 3 minutes. The amplified product was visualized on a 1.5% agarose gel, which was stained with ethidium bromide and run at 70 volts for 30 minutes. The fragment size was compared to a 100-bp molecular weight marker. Both sides of the amplified DNA fragment were sequenced, and the editing and alignment were performed using Sequencer software (version 6.1.0, Gene Codes Corp.).
For the phylogenetic analysis of HV,the obtained sequences were compared with 560 mtDNA control region sequences deposited in GenBank, which represent 50 animals of the Bos taurus and Bos indicus breeds.
Statistical analysis
To infer the spatial distribution of haplotypes, a nested clade analysis was performed using TCS software (version 1.1.3). This analysis represents the lowest possible number of steps (parsimonious method), which is interpreted as a distance measure, to accommodate a haplotype network (Clement et al., 2000). The distribution of HV haplotypes and the relationships with the Colombian Creole, South American, European, Indian and African breeds were determined by constructing median-joining networks for the haplotypes (Bandelt et al., 1999) using NETWORK software (version 4.1.0.8, Fluxus Technology Ltd., 2004).
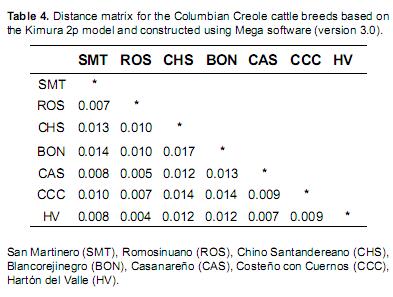
Polymorphisms were identified by comparing the sequences to the reference sequence NC_001567 (2), which corresponds to the central haplotype (Eucons) of the European taurine cluster (T3) (Bandelt et al., 1999). Variations from the consensus sequence or from another root haplotype (Afcons) were recorded. The Afcons haplotype is considered the root of the African taurine cluster (T1) and differs from Eucons by the C→T, T→C and T→C transitions at positions 16050, 16113 and 16255, respectively, which can be written as Afcons (Eucons050T-113C-255C). Distance matrices were generated according to the Kimura 2p model. Consensus networks were taken into account for the phylogenetic trees, which were constructed using the neighbor-joining method and Mega software (version 3.0) (Kumar et al., 2005).
Results
Diversity of the mtDNA control region
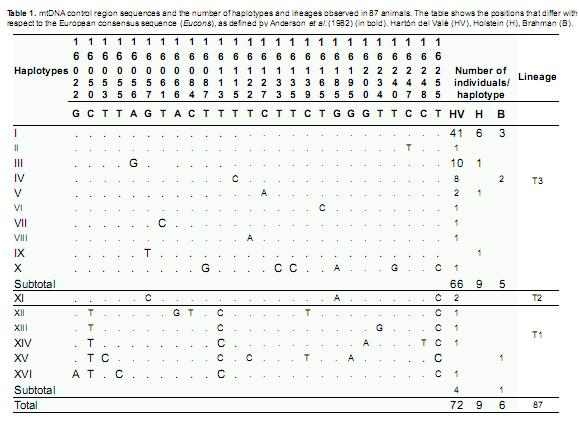
Haplotypes and lineages: The analysis of the mtDNA hyper-variable region in 72 sequences from HV cattle and in 15 sequences from the reference breeds revealed 16 mitochondrial haplotypes defined by the polymorphisms at 26 sites (Table 1). In HV, 14 haplotypes were identified based on 24 polymorphic sites (18 transitions and six transversions). Haplotypes I to X corresponded to the T3 lineage, which is characteristic of European breeds;haplotype XI was the only representative of the T2 lineage, which is characteristic of Middle Eastern breeds; and haplotypes XII to XVI corresponded to the T1 lineage, which is characteristic of African breeds.Haplotype XV was unique to B, and haplotype IX was unique to H. All H individuals corresponded to the T3 lineage.
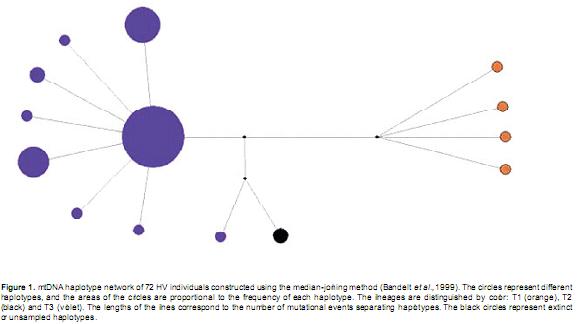
Zebu haplotypes were not observed in B, and 83% of the individuals belonged to the European lineage. The haplotype unique to B, haplotype XV, is derived from Afcons and has only been reported in Latin American Creole breeds; this haplotype was not observed in HV. In accordance with its European ancestry, the most common lineage in HV was the T3 lineage (91.7%), which was represented by nine haplotypes (Table 1, Figure 1). There were four T1 haplotypes (5.5%), indicating the contribution of African breeds to HV, and there was a single haplotype for the Near Eastern T2 lineage (2.7%). The most frequent haplotypes were I (57%), III (14%) and IV (11%) of the T3 lineage. The distribution of the haplotypes showed three levels of aggregation: the nine haplotypes of the T3 lineage, haplotype X of the T3 and T2 lineages and the four haplotypes of the African T1 lineage.
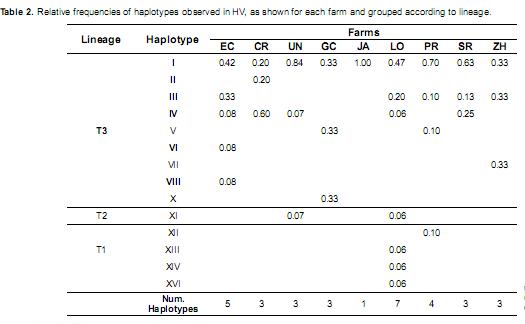
Among the nine farms, the LO estate had the only cattle population in which three lineages were observed, and there were seven total haplotypes in LO (Table 2). The UN and PR farms each had two lineages, and all other farms represented only the T3 lineage. Haplotype I was present in all of the sampled populations, and all of the JA individuals belonged to this group. Haplotypes III and IV were found in five and four populations, respectively, and haplotypes II, VI and VII were unique to CR, EC and ZH, respectively.
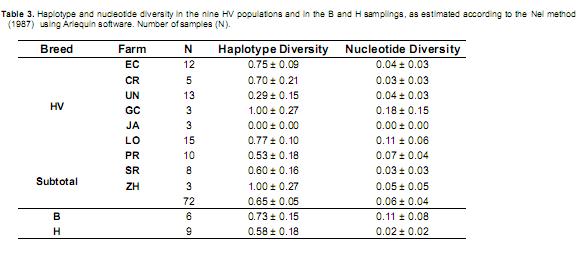
The average haplotype diversity was estimated using the Nei method (1987). In the nine HV herds, the diversity was 0.65 ± 0.05 (Table 3), which was lower than the diversity observed in B but greater than that in H. The GC, ZH and LO herds were notable for their high levels of haplotype diversity, UN was notable for its low diversity (0.26) and JA was notable for its uniform mtDNA.
Molecular analysis of variance did not reveal significant differences between the genetic groups (HV, H and B), and the fixation index (FST)was not significant because the heterogeneity of variances did not allow for the detection of differentiation. A low genetic structure was observed among the HV farms(FST = 0.035, p>0.06). Significant differences were observed between EC and ZH, between CR and ZH and between LO and ZH.
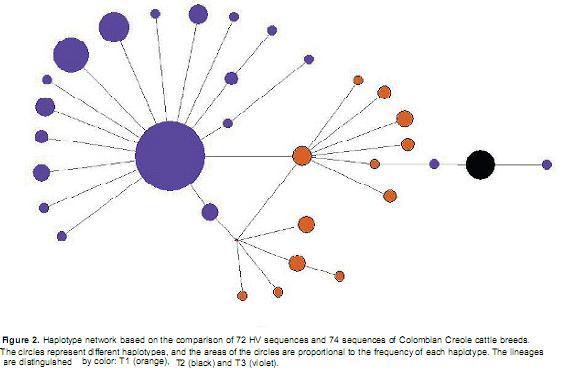
Relationship of the Hartón del Valle breed withother Colombian Creole cattle breeds. The 72 HV sequences were aligned with 74 sequences of Colombian Creole cattle breeds (CCB) available in GenBank (Carvajal-Carmona et al., 2003). The sequence comparisons revealed 19 haplotypes for the T3 lineage, 12 haplotypes for the T1 lineage and a single haplotype for the T2 lineage (Figure 2). Haplotypes V, VI, VII, VIII, X, XII and XIII identified in HV have not been reported in CCB. In accordance with the European ancestry of HV, most of the haplotypes corresponded to the T3 line. Of the 12 T1 haplotypes, four were observed in HV.
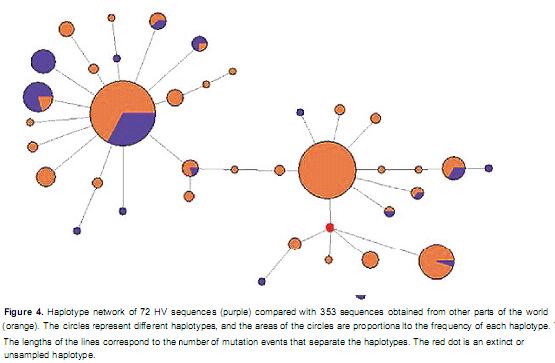
The relationship of the Hartón del Valle breed with other breeds from around the world. Using TCS software, a haplotype network was constructed with 560 sequences representing 50 Bos taurus and Bos indicus breeds from various regions. After validating the network and excluding unrelated sequences from the information generated in this study, 353 sequences remained, which represented only the Iberian, African and Latin American Creole breeds. The network revealed the presence of 37 different haplotypes; nine of the haplotypes were observed in HV, and seven were unique (Figure 4). Fifty percent of the sequences (175/353) represented the central haplotype of the T3 cluster (Eucons). This group included 87 HV sequences, 29 sequences from Spanish breeds (Pajuna, Lidia, RubiaGallega, Avileña,Mostrenca,Murciana, Monchina, Morucha, Negra Serrana, Retinta and Tudanca), 15 sequences from Portuguese breeds (Preta,Maronesa, Garvonesa and Mertolenga), 10 sequences from Argentine Creole breeds, 2 sequences from Bolivian Creole breeds, 6 sequences from Caribbean Creole breeds and 3 sequences from the African Kuri breed. Fifteen percent of the sequences represented the central haplotype of the African T1 cluster (Afcons), and the majority of these sequences (60%) came from animals of African cattle breeds (Egipcia, Kapsiki, N'Dama, Somba, White Fulani, Kuri and Kenana). A smaller number of the T1 sequences came from animals of Spanish, Portuguese, Caribbean Creole and Colombian Creole (one HV, two Romosinuano) breeds. The four HV sequences corresponding to the T1 lineage in this study were located in peripheral haplotypes. The unique haplotype representing the T2 lineage was present in five HV animals, two Blancorejinegro cattle and one Chino Santandereano individual. This T2 haplotype may have evolved in Colombia because it was not found in other breeds.
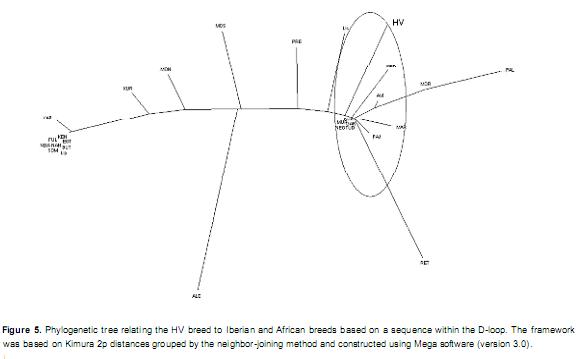
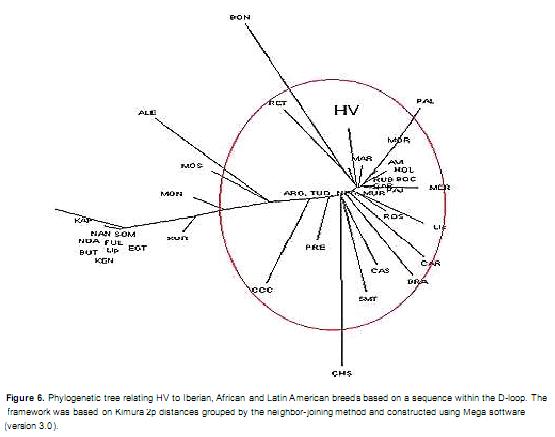
The relationship between HV and the Iberian and African breeds is shown in Figure 5. HV was found to exhibit phylogenetic proximity with the Iberian breeds, namely, the Spanish breeds of Tudanca, Rubia Gallega, Negra Serrana, Murciana, Pajuna and Avileña, and the Portuguese breeds of Garvonesa and Mertolenga. The African breeds were present in a different cluster, with the Kuri breed being the most closely related to the Iberian breeds and HV. A phylogenetic tree was constructed using the sequences from Iberian, African and Latin American breeds (Figure 6). The observed relationships confirmed the Iberian ancestry of and African influence on the Latin American Creole breeds; of these, only the Guadeloupe Creole breed was distant from the others. With the exception of the Blancorejinegro and Chino Santandereano breeds, the Colombian breeds were grouped into a cluster containing Spanish and Portuguese breeds along with the Creole breeds of Argentina, Bolivia and the Caribbean.
Discussion
Variation of the mtDNA control region in theHartón del Valle breed
The findings of the present study reveal the highly diverse collection of mtDNA in the HV breed; morespecifically, 14 haplotypeswere identified representing three mitochondrial lineages. The haplotypes were defined based on the polymorphisms at 24 sites. Eighteen of these polymorphisms were transitions, and six were transversions, thereby corroborating the transitional bias previously described in cattle (Bradley et al., 1996; Carvajal-Carmona et al., 2003; Loftus et al., 1994a). The number of haplotypes was greater than the numbers previously reported in HV (Carvajal- Carmona et al., 2003) and in four Brazilian Creole breeds and one Argentine breed (Miretti et al., 2002) and similar to the number identified in three Caribbean Creole breeds and the Nellore breed (Magee et al.,2002).
Cattle were introduced to the Caribbean by the Spanish conquistadors in 1493, and by 1525, cattle populations had expanded to Central and South America. Cattle shipments from Portugal to Brazil have also been documented (Primo, 1992), along with the introduction of African cattle to the continent during the sixteenth and seventeenth centuries as a result of the slave trade. As previously reported by Carvajal-Carmona et al. (2003), HV sequences represent three of the four Old World cattle lineages, defined by Troy et al. (2001). The findings of the present study reveal a large proportion of European mtDNA (T3) and a low proportion of African mtDNA (T1) within the HV breed. The distribution of the lineages was 5.5%, 2.7% and 91.7% for the T1, T2 and T3 lineages, respectively. Several studies have shown that T3 is the most common haplogroup among American Creole breeds (Lirón et al., 2006; Mirol et al., 2003; Miretti et al., 2002; Ginja et al., 2009).
These findings contrast sharply with the distribution previously reported for HV (38%, 29% and 33% for T1, T2 and T3, respectively) by Carvajal-Carmona et al. (2003). The difference may reflect the increased sample size in the present study (72 versus 21 samples), along with the different sizes and varied production systems of the farms included in the study.
Haplotypes of African origin have been detected in Portuguese and Iberian breeds (Beja-Pereira et al., 2006; Cymbron et al., 1999; Miretti et al., 2002), as well as American Creole breeds (Beja- Pereira et al., 2006; Egito et al., 2007; Lirón et al., 2008; Miretti et al., 2002; Mirol et al., 2003). A high maternal contribution of African breeds has been observed in Brazilian Creole breeds (T3=44.4% and T1=40.7%), and Brazil is the only South American country in which cattle of Portuguese origin contributed to the Creole breeds (Egito et al., 2007).
From 454 sequences of Creole (Colombia, Argentina, Brazil and West Indies), Iberian and African breeds, Lirón et al. (2006) found that the T3 haplogroup was the most common haplogroup among American Creole breeds (63.6%) followed by the African T1 haplogroup (32.4%) and the Middle Eastern T2 haplogroup (4%). Within the African lineage, two subclusters were identified: T1a, which has been reported in cattle in Brazil and the Lesser Antilles and with a distribution that does not include Africa, and T1, which is limited to the region of America colonized by the Spanish. In the present study, the majority of the HV samples corresponded to the European haplogroup (91.7%), and the percentage corresponding to the African lineage was significantly lower. A sample from a B individual corresponded the T1a haplotype, which may have originated in Brazil based on the findings that T1a is restricted to Brazil and the non-Spanish Caribbean Islands (Lirón et al., 2008). Two HV individuals belonging to the same population were of the T2 lineage, which is associated with breeds from the Near East and has only been reported in Colombia.
Several explanations have been proposed for the presence of African haplotypes in American Creole cattle: 1) the introduction of the T1 lineage to America from West Africa through intermediate slave trade ports, 2) the entry of African cattle onto the Iberian Peninsula during the Arab invasion prior to the conquest of America and 3) the contribution of Brazilian cattle with Portuguese ancestors that have African matrilineages (Lirón et al., 2008; Magee et al., 2002).
Haplotype diversity in HV (0.65 ± 0.05) was lower than that of breed B but greater than that of H. It is possible that the high diversity of HV has arisen since the time of the conquest and that a founding population of animals from different origins contributed to the breed. In support of this hypothesis, it has been reported that cattle arrived in the Valle del Cauca from the four cardinal directions (Finch, 1991). Cattle coming from the north and east derived from ancestral stock on the Caribbean coast, and those from the south originated in Ecuador. In 1540, cattle arrived in Cali via the trail through Dagua (further west); these cattle originated from Hispaniola and Nicaragua, and were introduced via the Panama Canal and the city of Buenaventura (Finch, 1991).
Among the nine farms, GC, ZH and LO had the highest haplotype diversity. LO was the only farm in which three mitochondrial lineages were observed, but the herd was liquidated in June 2009. The LO estate was established in 1890 and had the oldest tradition of cattle breeding, with founding animals that were descendents of the oldest herds in Roldanillo (Valle del Cauca). The high level of genetic diversity resulted from the use of a broad genetic base of pure livestock and the use of breeding bulls from other farms (Valderrama, 2008). The animals belonging to this herd were of great interest for the conservation of the genetic diversity of the breed, which was also observed using autosomal microsatellites and milk proteins (Álvarez, 2008).
Another herd with high diversity was GC, which, together with EC, belonged to Atogan Ltd., a cattle conglomerate dedicated to the production of milk and dairy products that was liquidated in 2008. The farms were established in 1955 with 40 HV cows, two HV bulls, two young HV-European mixed breeding bulls and 40 crossbred cows (Pardo Suizo, Zebu and Jersey). By eliminating the crossbreeds and by using the HV bulls, this conglomerate produced a purebred cattle herd. Later crosses were made using Holsteins and, to a lesser extent, German Anglers in order to improve milk production and certain characteristics of the breed.
By 1991, the farm had approximately 1,200 milking cows and, using artificial insemination, achieved anaverage production of 2,500kg milk/day (Valderrama, 2008). The artificial insemination technique used in this population favored the sampling of a larger number of bulls over natural mating. Therefore, the high diversity of this population was a result of a multiracial founder population and the widespread use of bulls and crossing events. The ZH farm, with origins that are unknown, exhibited high haplotype diversity but not nucleotide diversity; this observation most likely reflects the population size (1,000 animals) because no animals from other farms have been introduced into this population. The UN herd represented two mitochondrial lineages but exhibited low haplotype diversity (0.29 ± 0.15). This population originated from a mixed Holstein herd, and in 1968, animals from three different farms (which are no longer in existence) were introduced into the herd.
In 2004, UN received animals on loan from the Ministry of Agriculture and Development of the Valle del Cauca, with animals that had been inbred for 20 years and later included individuals from four other herds (Valderrama, 2008). The results of the present study provide information critical for increasing the genetic variation of the UN conservation herd at the National University of Colombia. The university is the only state entity that maintains the HV breed, which was not included in the programs established in the 1930s by the Colombian government for the protection of Creole cattle breeds or in the National Development Plan for Creole Cattle Breeds developed by the Ministry of Agriculture and Rural Development in 2004.
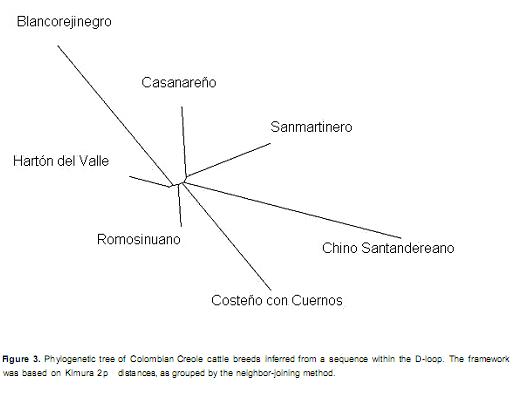
Relationship of the Hartón del Valle breed with Colombian and Creole cattle breeds. A comparison of the seven Colombian Creole breeds studied by Carvajal-Carmona et al. (2003) with those in the present study revealed a high degree of proximity between the HV and the Romosinuano breed and a lesser degree of proximity with the Costeño con Cuernos, Casanareño and San Martinero breeds. This proximity was previously reported by Pinzón (1984), who attributed the origin of Romosinuano, HV and Chino Santandereano to the Costeño con Cuernos. The study also described how cattle arrived at the Valle del Cauca from the four cardinal directions and how the cattle that came from the north and east were derived from ancestral stock developed on the Caribbean coast (Pinzón, 1991).
The Brahman breed was slightly more distant from HV, and it clustered with the Casanareño and San Martinero breeds (data not shown). Previous studies have revealed introgression with the Zebu breed in the Casanareño breed based on mtDNA analysis (Martínez et al., 2006) and cytogenetic studies of the Y chromosome (Sánchez et al., 2008).
Several studies have shown an early bifurcation in the bovine mtDNA between the two major taxa of domestic cattle, namely, Bos indicus and Bos taurus, indicating a divergence prior to domestication (Bradley et al., 1996; Loftus et al., 1994b). As in other reports investigating Bos indicus cattle in America, Zebu haplotypes were not observed in any of the Brahman samples analyzed. The American Zebu haplotypes were of taurine origin, indicating that they developed exclusively from female Bos taurus and male Bos indicus (Barrera et al., 2006; Egito et al., 2007; Meirelle et al., 1999; Mirol et al., 2003). In the United States, the origin of Brahman cattle dates back to the nineteenth century with contributions from the Guzerá, Nellore, Krishna Valley and Gyr breeds, which were imported from India and Brazil. In Colombia, Brahman cattle were crossed with Colombian Creole breeds and quickly spread throughout the country; this breed now constitutes more than 95% of the national Zebu herd (Pinzón 1991).
Relationship of the Hartón del Valle breed with other breeds worldwide
Although the identity of the ancestral founders of the American Creole breeds is uncertain, several reports have indicated the possible contribution of cattle from southern Spain. Some authors have claimed that ships came from Galicia in northern Spain and from the Canary Islands, a typical stopover for trips to America (Primo, 1992). Rouse (1977) discussed how the Spanish breeds Retinta, Berrenda, Cacereña and Andaluza Negra may have contributed to the development of American cattle. Casas and Valderrama (1998) suggested that the HV breed represents combinations of the following: Rubia Gallega and its descendants, Palmeña and Canaria; Asturiana de los Valles; and Menorquina, or Mahonesa. In the present study, HV was found to have high proximity with Iberian breeds and low proximity with African breeds. The closest Iberian breeds included the following: Tudanca, Rubia Gallega, Negra Serrana, Murciana, Pajuna and Avileña, as well as the Portuguese breeds Garvonesa and Mertolenga.
The study provides information regarding the conservation of the Hartón del Valle breed, whichhasadecliningpopulation. Thegene pool of Hartón del Valle represents at least three sources: an African origin (5.5%), a Mediterranean European origin from the Middle East (2.7%) and a Mediterranean European origin (91.7%).
Acknowledgments
This work was supported by the Research Division of the National University of Colombia in Palmira (DIPAL). We wish to acknowledge the producers of Hartón del Valle cattle for their contribution of samples, especially Dr. Marino Valderrama, Dr. Eduaime Cárdenas and Dr. Germán Ortiz. We thank Andrea Egito of Embrapa (Cenargen-Brazil) for her constant advice and the biologist Andrés M. Posso of the National University of Colombia at Palmira. We also thank the Center for Tropical Agriculture (CIAT) for assistance with data processing, and we thank the biologist Diana López of the Alexander von Humboldt Institute.
References
1. Álvarez LA. Genetic diversity of Hartón del Valle cattle and their relationship with Holstein and Brahman through the use of molecular markers. Doctoral Dissertation, Faculty of Science, University of Valle, Colombia Cali 2008. [ Links ]
2. Anderson S, de Bruijn MH, Coulson AR, Eperon C, Sanger F, Young IG. Complete sequence of bovine mitochondrial DNA: conserved features of the mammalian mitochondrial genome. J Mol Biol 1982; 156:683-717. [ Links ]
3. Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 16:37-48. [ Links ]
4. Barrera G, Martínez R, Pérez JE, Herazo T. Identification of Bos Taurus mitochondrial DNA in populations of Colombian Zebu Brahman cattle. Rev Corpoica - Agricultural Science and Technology 2006; 7:21-24. [ Links ]
5. Beja-Pereira A, Caramelli D, Lalueza-Fox C, Vernesi C, Ferrand N, Casoli F, Goyache LJ, Royo S, Conti M, Lari A, Martini L, Ouragh A, Magid A, Atash A, Zsolnai P, Boscato C, Triantaphylidis K, Ploumi KL, Sineo F, Mallegni P, Taberlet G, Erhardt L, Sampietro J, Bertranpetit G, Barbujani G, Luikart and Bertorello G. The origin of European cattle: Evidence from modern and ancient DNA. Proc Natl Acad Sci 2006; 103:8113-8118. [ Links ]
6. Bradley DG, MacHugh DE, Cunningham P and Loftus RT. Mitochondrial diversity and the origins of African and European cattle. Proc Natl Acad Sci 1996; 93:5131-5135. [ Links ]
7. Bruford MW, Bradley DG, Luikart G. DNA markers reveal the complexity of livestock domestication. Nat Rev Genet 2003; 4:900-910. [ Links ]
8. Casas I, Valderrama M. The Hartón del Valle Creole Cattle Breed. National University of Colombia at Palmira; 1998. [ Links ]
9. Carvajal-Carmona L, Bermudez N, Olivera-ingel M, Estrada L, Ossa J, Bedoya G, Ruiz-Linares A. Abundant mtDNA Diversity and Ancestral Admixture in Colombian criollo Cattle (Bostaurus) Genet 2003; 165:1457-1463. [ Links ]
10. Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Mol Ecol 2000; 9:1657-1660. [ Links ]
11. Cymbron T, Ronan T, Loftus RT, Malhiero M, Bradley D. Mitochondrial sequence variation suggests an African Influence in Portuguese cattle. Proc R Soc Lond 1999; 266:597-603. [ Links ]
12. Egito A. Genetic diversity, individual ancestry and interbreeding in Brazilian cattle breeds based on microsatellite analysis and mtDNA haplotypes:subsidies for conservation. Doctoral Dissertation, University of Brasilía, Brazil 2007. [ Links ]
13. Fluxus Technology Ltd, NetWork program version 4.1.0.8 2004 [Date accessed: June 2008] http://www.fluxus-engineering.com [ Links ]
14. Ginja C, Melucci L, Quiroz J, Revidatti MA, Martínez- Martínez A, Delgado JV, Penedo MCT, Gama LT. Origin and genetic diversity of New World Creole cattle: inferences from mitochondrial and Y chromosome polymorphisms. Anim Genet 2009; 41:128-41. [ Links ]
15. Kumar S, Tamura K, Nei M. MEGA3: Integrated Software for Molecular Evolutionary Genetics Analysis and Sequence Alignment. Brief Bioinform 2005; 5: 150-163. [ Links ]
16. Lirón JP, Bravi CM, Mirol PM, Peral-García P, Giovambattista G. African matrilineages in American Creole cattle: evidence of two independent continental sources. Anim Genet 2006; 37:379-382. [ Links ]
17. Lirón JP, Diego MP, Michael D, MacNeil PM, Mirol AR, Rogberg-Muñoz A, Martínez-Martínez A, Maciel S, Terán-Polo C, Peral-García P, Hanotte O, Giovambattista G. Historical origin andphylogeographicaffinity of American Creole cattle through the study of mitochondrial DNA. IX Latin American symposium on the conservation and utilization of animal genetic resources. Buenos Aires (Argentina) National University of Lomas de Zamora. 2008; p. 311-316. [ Links ]
18. Loftus RT, MacHugh DE, Bradley DG, Sharp PM, Cunningham EP. Evidence for two independent domestications of cattle. Proc Natl Acad Sci 1994a; 91:2757-2761. [ Links ]
19. Loftus RT, MacHugh DE, Ngere LO, Balain DS, Badi AM, Bradley DG, Cunningham EP. Mitochondrial genetic variation in European, African and Indian cattle populations. Anim Genet 1994b; 25:265-271. [ Links ]
20. Magee DA, Meghen C, Harrison S, Trio C, Cymbron T, Gaillard C, Morrow A, Maillard J, Bradley D. A Partial African Ancestry for the Creole Cattle Populations of the Caribbean. J Hered 2002; 93:429-432. [ Links ]
21. Martínez G. Editor. Census and characterization of Colombian Creole cattle production systems. National Cattle Fund, ICA, PRONATTA, Asobón, 1999. [ Links ]
22. Martínez R, Barrera G, Sastre H. Variability and genetic status of seven subpopulations of the Casanareño Colombian Creole breed. Rev Corpoica 2006; 7:5-11 [ Links ]
23. Meirelle F, Rosa A, Lobo R, Smith J, Duarte F. Is the American Zebu really Bosindicus? Genet Mol Biol 1999; 22:543-546. [ Links ]
24. Miller SA, Dikes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res 1988; 16:1215. [ Links ]
25. Miretti MM, Pereira HA, Poli MA, Contel EP, Ferro JA. African-derived mitochondria in South American native cattle breeds (Bos taurus): evidence of a new taurine mitochondrial lineage. J Hered 2002; 93:323-330. [ Links ]
26. Mirol PM, Giovambattista G, Lirón JP, Dulout FN. African and European mitochondrial haplotypes in South American Creole cattle. Hered 2003; 91:248-254. [ Links ]
27. Nei M. Molecular Evolutionary Genetics. Columbia University Press, (NY), USA, 1987. [ Links ]
28. Pinzón E. History of cattle in Colombia. Ed. 1 (Colombia): Cattle Bank, 1984. [ Links ]
29. Pinzón E. History of cattle in Colombia. (Colombia): Cattle Bank, 1991. [ Links ]
30. Primo A. Iberian cattle in the Americas: 500 years later. Arch Zootec 1992; 41: 421-432. [ Links ]
31. Rouse JE. The Creole: Spanish cattle in the Americas. NORMAN. Oklahoma: University of Oklahoma Press, USA, 1977. [ Links ]
32. Sánchez CA, JimÈnez L, Bueno M. Genetic introgression of Bos indicus (Bovidae) in Colombian Creole cattle of Bos taurus origin. Acta Biol Colomb 2008; 13:131-142. [ Links ]
33. Troy CS, MacHugh DE, Bailey JF, Magee DA, Loftus RT, Cunningham P, Chamberlain AT, Sykes BC, Bradley DG. Genetic Evidence for Near-Eastern origins of European cattle. Nature 2001; 410:1088-1091. [ Links ]
34. Valderrama M. Hartón del Valle. Creole dairy cattle of the Cauca River Valley. 2008. [ Links ]