Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Colombiana de Ciencias Pecuarias
Print version ISSN 0120-0690
Rev Colom Cienc Pecua vol.26 no.4 Medellín Oct./Dec. 2013
ORIGINAL ARTICLES
Stearoyl CoA desaturase and fatty acid synthase gene polymorphisms and milk fatty acid composition in Chilean Black Friesian cows¤
Polimorfismos en genes Estearoyl CoA desaturasa y ácido graso sintasa y composición de ácidos grasos en la leche de las vacas Frisón Negro Chileno
Polimorfismos no gene Estearoyl CoA desaturase e ácido graxo sintase e composição de ácidos graxos do leite de vacas Frison Preto Chileno
Karla B Inostroza*1, PhD; Erick S Scheuermann2, PhD; Néstor A Sepúlveda3, MV, PhD.
* Corresponding author: Karla B Inostroza, Universidad de La Frontera, Avenida Francisco Salazar N° 01145, Temuco, Chile. Email: karla.inostroza@gmail.com
1Laboratorio de Producción Animal, Facultad de Ciencias Agropecuarias y Forestales, Universidad de La Frontera, Temuco, Chile.
2Departamento de Ingeniería Química, Universidad de La Frontera, Temuco, Chile.
3Facultad de Ciencias Agropecuarias y Forestales, Universidad de La Frontera, Temuco, Chile.
(Received: June 14, 2012; accepted: October 31, 2012)
Summary
Background: in recent years, market attention has increasingly focused on improving the quality of dairy products. Therefore, animal selection for healthier milk fatty acid composition are therefore of interest. Objective: the aim of this study was to identify whether two polymorphisms (c.878T>C in the SCD gene and g.17924A>G in the FASN gene) are related with the fatty acid composition of milk. Methods: individual milk samples were collected from 50 Chilean Black Friesian cows. Fatty acids (FA) from each milk sample were extracted, analyzed, and quantified (mg FA/g of milk) by gas chromatography. The genotype of each sample was also determined using PCR-RFLP (Polymerase Chain Reaction – Restriction Fragment Length Polymorphism). Results: milk from cows with a c.878CC genotype in the SCD gene exhibited a greater content of C14:1, C17:1, C18:1n9t, C18:1n9c, and total monounsaturated fatty acids (MUFA), as well as a higher 14:1/14 ratio than other SCD genotypes. In addition, considering the importance of the predominant isomer of conjugated linoleic acid for human diet, we identified a higher content of cis-9, trans-11 conjugated linoleic acid in milk samples from animals that had the c.878CC genotype. For the FASN gene, cows with the g.17924GG genotype presented high contents of C14:0, C16:0, C18:0, C18:1n9t, C18:1n9c, and MUFA. Conclusion: therefore, these polymorphisms could be useful genetic markers for studying the fatty acid composition of dairy milk.
Key words: conjugated linoleic acid, monounsaturated, SNP.
Resumen
Antecedentes: en los últimos años, la atención del mercado se ha centrado cada vez más en mejorar la calidad los productos lácteos. Por lo tanto, la selección de animales con una composición de ácidos grasos de leche más saludable es de interés. Objetivo: el principal objetivo de este estudio fue identificar dos polimorfismos (c.878T>C en el gen SCD y g.17924A>G en el gen FASN) y su relación con la composición de ácidos grasos de leche. Métodos: muestras individuales de leche fueron recolectadas de 50 vacas Frison Negro Chileno. Los ácidos grasos (FA) de cada muestra de leche fueron extraídos, analizados y cuantificados (mg FA/g leche) por cromatografía de gases. El genotipo de cada muestra se determinó mediante la técnica PCR-RFLP (reacción en cadena de la polimerasa - polimorfismo de longitud de fragmentos de restricción). Resultados: los animales con el genotipo c.878CC en el gen SCD presentaron un mayor contenido de C14:1, C17:1, C18:1n9t, C18:1n9c y total de ácidos grasos monoinsaturados (MUFA), así como elevados índices de 14:1/14 que otros genotipos SCD. Además, debido a la importancia en la dieta humana del isómero predominante de ácido linoleico conjugado (CLA), hemos identificado un mayor contenido de cis-9, trans-11 CLA en muestras de leche de animales que poseen el genotipo c.878CC. Para el gen FASN, vacas con el genotipo g.17924GG presentaron un mayor contenido de C14:0, C16:0, C18:0, C18:1n9t, C18:1n9c y contenido total de ácidos grasos monoinsaturados (MUFA). Conclusión: por lo tanto, estos polimorfismos podrían servir como marcadores genéticos para el estudio de la composición de ácidos grasos en la leche de vaca.
Palabras clave: ácido linoleico conjugado, monoinsaturados, SNP.
Resumo
Antecedentes: nos últimos anos, o mercado fornecedor de laticínios tem-se voltado ao melhoramento da qualidade dos seus produtos. Por conseguinte, é de interesse a seleção de animais que apresentem uma composição de ácidos graxos mais saudáveis no seu leite. Objetivo: o objetivo principal deste estudo foi identificar dois polimorfismos (c.878T>C no gene SCD e g.17924A>G no gene FASN) e sua associação com a composição de ácidos graxos do leite. Métodos: as amostras de leite foram coletadas de 50 vacas Frison Preto Chileno. Os ácidos graxos (FA) de cada amostra de leite foram extraídos, analisados e quantificados (mg FA/g leite) por cromatografia gasosa. O genótipo de cada amostra foi determinado pela reação de PCR-RFLP (Reação em cadeia de polimerase - polimorfismo de comprimento dos fragmentos de restrição). Resultados: os animais com o genótipo c.878CC no gene SCD têm um teor mais elevado de C14:1, C17:1, C18:1n9t, C18:1n9c e do total de ácidos graxos monoinsaturados (AGM), além de níveis mais elevados de 14:1/14 que outros genótipos SCD. Além disto, devido à importância na dieta humana do isômero predominante do ácido linoléico conjugado (CLA), identificou-se um maior teor de cis-9, trans-11 CLA nas amostras de leite de animais que tinham o genótipo c.878CC. Para o gene FASN, as vacas com o genótipo g.17924GG tinham um teor mais elevado de C14:0, C16:0, C18:0, C18:1n9t, C18:1n9c e AGM. Conclusão: estes polimorfismos poderiam ser usados como marcadores genéticos para o estudo da composição de ácidos graxos do leite de vaca.
Palavras chave: ácido linoléico conjugado, monoinsaturados, SNP.
Introduction
Milk is a natural source of conjugated linoleic acid (CLA), which is a mixture of positional and geometric isomers of linoleic acid, with positive effects on human health (Pariza et al., 2000; Barceló-Coblijn and Murphy, 2009). The anticancer activity of CLA has been clearly established in cell culture and in animal models for a wide range of cancers (Lock and Bauman, 2004). The predominant source of CLA in human diets is derived from ruminant food products, with dairy products contributing nearly 75% of the total in human diet (Ritzenthaler et al., 2001; Parodi, 2003).
Several studies indicate that the average proportions of fatty acids in bovine milk are 70% saturated, 25% monounsaturated, and 5% polyunsaturated fatty acids (Grummer, 1991; Jensen, 2002; Kalac and Samkova, 2010). This composition is determined by factors related to food and to the bovine genotype, indicating that the selection of livestock can be an effective tool for improving milk fat composition (Schennink et al., 2008). A method that would accelerate selection of dairy cows with high concentration of beneficial fatty acids consists in identifying the genetic markers and/or polymorphisms associated with milk fat composition (Moioli et al., 2007).
Several enzymes affect lipid metabolism in the mammary gland. Stearoyl-CoA desaturase (SCD) is responsible for converting saturated fatty acids into monounsaturated fatty acids (Taniguchi et al., 2004). The SCD gene has also been suggested to be a major gene for changing the ratio of unsaturated to saturated fatty acids in milk and increasing CLA content (Moioli et al., 2007). Overall, 70% of cis- 9, trans-11 CLA in ruminant milk is produced in the mammary tissue by SCD enzyme activity (Bauman et al., 2006). The SNP c.878T>C results in the incorporation of a different amino acid: alanine (allele C) or valine (allele T). The Val residue may change the enzyme's catalytic activity compared with Ala (Taniguchi et al., 2004). Fatty Acid Synthase (FASN) is part of a multifunctional enzyme complex that catalyzes the synthesis of saturated fatty acids, such as myristate, palmitate and stearate (Smith et al., 2003; Roy et al., 2006). A g.17924A>G polymorphism resides in the thioesterase domain (TE) of the FASN gene and is responsible for determining the length of FASN targets (Zhang et al., 2008). Therefore, TE can contribute to both the contents of C14:0 and C16:0 as well as the contents of other fatty acids.
The aim of this study was to determine if two single nucleotide polymorphisms (SNPs) found in either the SCD (c.878T>C) or FASN (g.17924A>G) genes are associated with the fatty acid composition of milk in Chilean Black Friesian cows.
Materials and methods
This research was approved by the Bioethics Committee of University of La Frontera, Chile (June 1, 2011).
SNP Determination
Milk samples were individually collected from 50 Chilean Black Friesian cows and frozen at -20 °C until analysis. The cows were kept in the same group during the study and received a diet based on grazing mixed pastures (rye-grass and white clover) supplemented with 6 kg per head daily of a commercial concentrate (17% crude protein, 12% crude fiber, 2.69 Mcal/kg dry matter as metabolic energy).
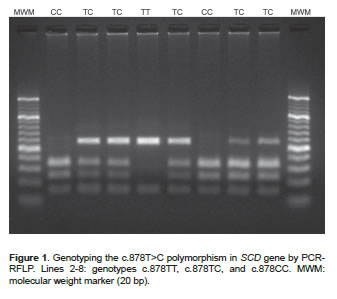
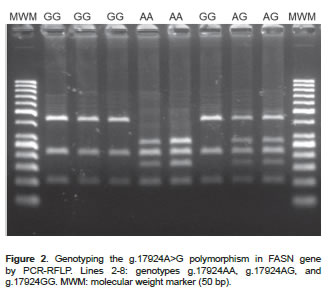
DNA was extracted from blood samples using AxyPrep Miniprep Kit (Axygen Biosciences, Union City, CA, USA). The presence of SNPs was evaluated by PCR-RFLP. Primers were designed with a Primer3 program based on the sequence available in GenBank (AF285607 and AY241932). A 170 bp region of the SCD gene was amplified through 30 amplification cycles (95 ºC/20 sec, 61 ºC/20 sec, and 72 ºC/30 sec) using the primers 5`-ACCTGGCTGGTGAATAGTGCT-3' and 5'-TCTGGCACGTAACCTAATACCCT-3'. A 758 bp region of the FASN gene was amplified through 30 amplification cycles (95ºC/20 sec, 58 ºC/20 sec, and 72 ºC/30 sec) using the primers 5'-TGCTGTCCATGCGGGAAATC-3' and 5'-TGAGCACATCTCGAAAGCCA-3'. SNPs of interest were then genotyped by RFLP using two enzymes: SatI (10U/μl, Fermentas Inc, Glen Burnie, MD, USA) and MscI (5U/μl, Fermentas Inc, Glen Burnie, MD, USA). After digestion (37 ºC for 4 hours), the restriction fragments were loaded onto a 3% agarose gel and separated by gel electrophoresis. The possible genotype patterns were as follows: 122, 28 and 20 bp (TT), 122, 75, 47, 28 and 20 bp (TC), and 75, 47, 28 and 20 bp (CC) for the SNP c.878T>C; or 273, 217, 167 and 101 bp (AA), 440, 273, 217, 167 and 101 bp (AG), and 440, 217, and 101 bp (GG) for the SNP g.17924A>G.
Fatty acids analysis
The Rose-Gottlieb method (AOAC, 1990) was used to extract the fatty acids from milk. The fatty acids obtained from milk were subjected to methylation with KOH 2N in methanol and hexane. Fatty acid methyl esters were then analyzed by gas chromatography (Clarus 500, Perkin Elmer, Buckinghamshire, UK) equipped with a flame ionization detector and a column Fused Silica Capillary Column SPTM 2380 (60m x 0.25mm x 0.2 μm film thickness) (Supelco, Bellefonte, PA, USA). One microliter was injected using the following temperature program: 150 ºC for 1 min followed by an increase of 1 °C/min to 168 ºC, held for 11 minutes, followed by an increase of 6 ºC/min to 230 °C, and then maintained for 8 minutes. Fatty acids were identified using the standard mixture of FAME Mix (C4-C24, Supelco, Bellefonte, PA, USA), and isomers of CLA were identified using Octadecadienoic Acid, Conjugated Methyl Ester (CLA Sigma-Aldrich, Milwaukee, WI, USA) as a standard. All fatty acids were quantified using calibration curves. Methyl Nonadecanoate (Sigma- Aldrich, Milwaukee, WI, USA) was used as an internal standard.
Statistical analysis
Data were analyzed using a general linear model in the SPSS version 17.0 for Windows (SPSS Inc, Chicago IL, USA). The equation can be represented as yijk= μ + Parityi + DIMj + GENk + εijk, where y is the dependent variable, μ= overall mean, Parityi= effect of the ith parity (1, 2, >3), DIMj = effect of jth days in milking interval (100, 100-200, >200), GENk = effect of kth genotype (SCD-FASN), and εijk = random residual. The results on the effects of the FASN and SCD genotype are presented as estimated marginal means ± SE (p<0.05).
Results
Genotypic and allele frequencies
The descriptive analysis of the polymorphisms indicates that the populations are at Hardy-Weinberg equilibrium. Genotypic and allele frequencies for the SNP c.878T>C were found to be TT: 14.4%, TC: 47.2%, CC: 38.4% and T=0.38 and C=0.62, respectively (Figure 1). For the SNP g.17924A>G the genotypic frequencies were AA: 9.6%, AG: 42.8, and GG: 47.6%, and the allele frequencies were A=0.31 and G=0.69 (Figure 2).
c.878T>C polymorphism in the SCD gene
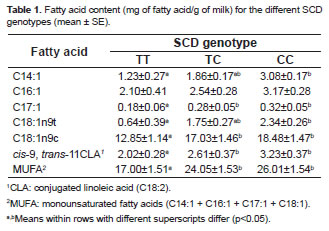
The c.878T>C SNP was related with the concentrations of several fatty acids. Table 1 shows the means, SE and SCD genotype comparisons for fatty acid composition in milk. After all fatty acids were analyzed, the animals with c.878CC genotype had higher values of myristoleic (C14:1), cis-10-Heptadecenoic (C17:1), elaidic (C18:1n9t) and oleic acid (C18:1n9c). In contrast, similar contents of palmitoleic acid (C16:1) were observed in the genotypes SCD. However, the total monounsaturated fatty acids (MUFA) content (C14:1 + C16:1 + C17:1 + C18:1) was greater in cows with genotype c.878CC (p<0.05). The cis-9, trans-11CLA content in the cows with the c.878CC genotype was greater than in those of the c.878TT genotype (p<0.05).
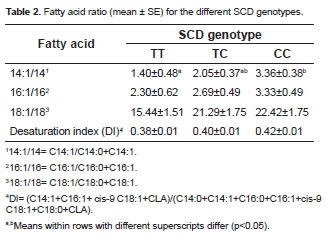
Table 2 shows the relationship of fatty acids, such as product and substrate of desaturation, for the different SCD genotypes. This analysis of desaturation activity indicators highlighted a significant effect on the SCD polymorphism only on the 14:1/14 ratio in c.878CC genotype.
FASN gene polymorphism g.17924A>G
It was predicted that the SNP g.17924A>G causes an amino acid replacement from threonine to alanine in the fatty acid synthase TE domain. In this study, the SNP was found to have relations with the major saturated fatty acids. The content of myristic (C14:0), palmitic (C16:0), and stearic acid (C18:0) were greater (p<0.05) in Chilean Friesian cows with the g.17924GG genotype than those with the g.17924AA genotype. The g.17924GG genotype cows had greater MUFA content than cows with g.17924AA genotype (Table 3).
Discussion
Genotypic and allele frequencies
Allele frequencies for the SNP c.878T>C were found to be T=0.38 and C=0.62. In Italian Red Pied cows C=0.34, and in Italian Grey Alpine cows, was C=0.89 (Milanesi et al., 2007), compared with the T allele, the higher frequency of the C allele is in agreement with result reported previously. For the SNP g.17924A>G, A=0.31 and G=0.69. In Friesian cows, the frequencies of the A allele were 0.53 (Schennink et al., 2009) and 0.31 (Morris et al., 2007), which are similar to the frequency found in our population.
c.878T>C polymorphism in the SCD gene
Polymorphism in the 5th exon (c.878TC) was predicted to result in an amino acid replacement from valine to alanine in the SCD protein (Taniguchi et al., 2004). The C allele has been associated with high SCD enzyme activity and concentration of monounsaturated fatty acids (MUFA) in milk (Milanesi et al., 2007). The differences obtained in the fatty acids C14:1, C17:1, C18:1n9t, C18:1n9c and MUFA coincide with results reported in the fatty acid composition of beef cattle (Taniguchi et al., 2004). The relation of the c.878CC genotype with a higher content of myristoleic acid (C14:1) was also found in brisket adipose tissue, however, no significant association were detected for palmitoleic (C16:1) and oleic acid (C18:1n9c) (Li et al., 2011). In this study, the association with palmitoleic acid was not significant. Li et al. (2011) suggested that other SNPs of SCD or other genes may also play a role in the desaturation of palmitate (C16:0) and stearate (C18:0). On the other hand, the different contents of cis-10 heptadecanoic fatty acid (C17:1) based on SCD genotypes has not been previously reported in studies related to this polymorphism. This result suggests that C17:0 could be affected by SCD activity.
In milk, the content of cis-9, trans-11 CLA exhibits individual variation that can be observed in cows fed the same ration (Bauman et al., 2001) suggesting an important role for genetics. In our study, the c.878CC genotype has a higher content of cis-9, trans-11 CLA compared with the c.878TT genotype.
Only statistically significant differences in the ratio 14:1/14 were found in the desaturation analysis, which agrees with the study by Mele et al. (2007) in Holstein cows. The ratio 14:1/14 has been suggested to be the best measure of SCD activity (Lock and Garnsworthy, 2003). The lack of a relationship between SCD genotypes and other fatty acids can be attributed to the wide range of factors that influence the activity of desaturation in the mammary gland. Moreover, C14:0 in milk is derived almost exclusively from de novo synthesis in the mammary gland. Therefore, most of the C14:1 is likely to be synthesized by SCD (Bernard et al., 2006).
FASN gene polymorphism g.17924A>G
The TE domain is located at the 3'-end of FASN and is encoded within four exons (exons 39-42). This domain is responsible for the determination of the length of FASN products, which are potential substrates for elongases and desaturase (Zhang et al., 2008). The g.17924AG SNP present in the TE domain of the FASN gene has been associated with the content of C14:0 in subcutaneous adipose tissue and milk fat (Morris et al., 2007). We found that FASN genotype significantly affected myristic (C14:0), palmitic (C16:0), stearic (C18:0), elaidic (C18:1n9t), and oleic acid (C18:1n9c). Elaidic (C18:1n9t) and oleic acid (C18:1n9c) differences in contents can be explained if more C16:0 is produced by FASN (genotype g.17924GG), then more C16:0 will be elongated to C18:0 that will then be converted to C18:1(n-9), which is the main product of fatty acid de novo synthesis (Zhang et al., 2008). The differences in MUFA can also be explained if more C14:0 and C16:0 were produced, as a result of the FASN g.17924GG genotype, that were consequently converted to more C14:1 and C16:1 by SCD. Therefore, both SCD and FASN genotypes contribute to the MUFA content.
SCD and FASN are two key genes involved in the pathway for MUFA synthesis (Kim and Ntambi, 1999). Li et al. (2011) examined the epistatic effect of the two-gene SNP genotypes on the fatty acid composition, and the results suggested that the role of the SCD in fatty acid desaturation may be modified in the presence of different genotypes of FASN and vice versa.
Therefore, we conclude that there is a relationship between the FASN and SCD genotypes with regards to the content of some fatty acids in Chilean Black Friesian cow milk. Our results suggest that the FASN and SCD genotypes can be a useful genetic tool to improve the nutritional quality of Chilean Black Friesian cow milk.
¤ To cite this article: Inostroza KB, Scheuermann ES, Sepúlveda NA. Stearoyl CoA desaturase and fatty acid synthase gene polymorphisms and milk fatty acid composition in Chilean Black Friesian cows. Rev Colomb Cienc Pecu 2013; 26:263-269.
Acknowledgements
We are grateful to the Comisión Nacional de Investigación Científica y Tecnológica (CONICYT Chile) for the financial support of this study.
References
Barceló-Coblijn G, Murphy EJ. Alpha linolenic acid and its conversion to longer chain n-3 fatty acid: benefits for human health and role in maintaining tissue n-3 fatty acid levels. Prog Lipid Res 2009; 48:355-374. [ Links ]
Bauman D, Corl B, Baumgard L, Griinari J. Conjugated Linoleic Acid (CLA) and the dairy cow. In: Garnsworthy P, Wiseman J, editors. Recent Advances in Animal Nutrition. Nottingham: Nottingham Univ. Press; 2001. p.221-250. [ Links ]
Bauman D, Mather I, Wall R, Lock L. Major advances associated with biosynthesis of milk. J Dairy Sci 2006; 89:1235-1243. [ Links ]
Bernard L, Leroux C, Chilliard Y. Characterization and nutritional regulation of the main genes in the lactating mammary gland. In: Sejrsen K, Hvelplund T, Nielsen MO, editors. Ruminant Physiology. Wagening: Wageningen Academy Publisher; 2006. p.295-326. [ Links ]
Grummer R. Effect of feed on the composition of milk fat. J Dairy Sci 1991; 74:3244-3257. [ Links ]
Jensen R. The composition of bovine milk lipids. J Dairy Sci 2002; 85:295-350. [ Links ]
Kalac P, Samkova E. The effects of feeding various forages on fatty acid composition of bovine milk fat: A review. Czech J Anim Sci 2010; 23:521-537. [ Links ]
Kim YC, Ntambi JM. Regulation of stearoyl-CoA desaturase genes: role in cellular metabolism and preadipocyte differentiation. Biochem Bioph Res Co 1999; 266:1-4. [ Links ]
Li C, Aldai N, Vinsky M, Dugan ME, McAllister TA. Association analyses of single nucleotide polymorphisms in bovine stearoyl-CoA desaturase and fatty acid synthase genes with fatty acid composition in commercial cross-bred beef steers. Anim Genet 2011; 43:93-97. [ Links ]
Lock A, Garnsworthy P. Seasonal variation in milk conjugated linoleic acid and Δ9-desaturase activity in dairy cows. Livest Prod Sci 2003; 79:47-59. [ Links ]
Lock A, Bauman D. Modifying milk fat composition of dairy cows to enhance fatty acids beneficial to human health. Lipids 2004; 39:1197-1206. [ Links ]
Mele M, Conte G, Castiglioni B, Chessa S, Macciotta N, Serra A, Buccioni A, Pagnacco G, Secchiari P. Stearoyl-Coenzyme A desaturase gene polymorphism and milk fatty acid composition in Italian Holstein. J Dairy Sci 2007; 90:4458-4465. [ Links ]
Milanesi E, Nicoloso L, Crepaldi P. Stearoyl CoA desaturase (SCD) gene polymorphisms in Italian cattle breeds. J Anim Breed Genet 2007; 125:63-67. [ Links ]
Moioli B, Contanari G, Avalli A, Catillo G, Orru L, De Matteis G, Masoero G, Napolitano F. Short communication: Effect of Stearoyl-Coenzyme A Desaturase polymorphism on fatty acid composition of milk. J Dairy Scie 2007; 90:3553-3558. [ Links ]
Morris C, Cullen N, Glass B, Hyndman D, Manley T, Hickey S, McEwan J, Pitchford W, Bottema C, Lee M. Fatty acid synthase effects on bovine adipose fat and milk fat. Mamm Genome 2007; 18:64-74. [ Links ]
Association of Official Analytical Chemists (AOAC). Official methods of analysis. In: AOAC, editors. AOAC. 15th ed. Washington: A.O.A.C. Press; 1990. p.10-18. [ Links ]
Pariza MW, Park Y, Cook M. Mechanism of action of conjugated linoleic acid: evidence and speculation. P Soc Exp Biol Med 2000; 223:8-13. [ Links ]
Parodi P. Conjugated linoleic acid in food. In: Sébédio J, Christie L, Adlof R, editors. Advances in Conjugated Linoleic Acid Research. Champaign: AOCS Press; 2003. p.101-122. [ Links ]
Ritzenthaler K, McGuire MK, Falen R, Shultz T, Dasgupta N, McGuire MA. Estimation of conjugated linoleic acid intake. J Nutr 2001; 131:1548-1554. [ Links ]
Roy R, Ordovas L, Zaragoza P, Romero A, Moreno C, Altarriba J, Rodellar C. Association of polymorphisms in the bovine FASN gen with milk-fat content. Anim Genet 2006; 37:215- 218. [ Links ]
Schennink A, Heck J, Bovenhuis H, Visker M, Van Valenberg H, Van Arendonk J. Milk fatty acid unsaturation: genetic parameters and effects of Stearoyl-CoA Desaturase (SCD) and Acyl CoA: Diacylglycerol Acyltransferase 1 (DGAT1). J Dairy Sci 2008; 91:2135-2143. [ Links ]
Schennink A, Bovenhuis H, Leon-Kloosterziel M, Van Arendonk M, Visker M. Effect of polymorphisms in the FASN, OLR1, PPARGC1A, PRL and STAT5A genes on bovine milkfat composition. Anim Genet 2009; 40:909-916. [ Links ]
Smith S, Witkowski A, Joshi A. Structural and functional organization of animal fatty acid synthase. Progr Lipid Res 2003; 42:289-317. [ Links ]
Taniguchi M, Utsugi T, Oyama K, Mannen H, Kobayashi M, Tanabe Y, Ogino A, Tsuji S. Genotype of stearoyl-CoA desaturase is associated with fatty acid composition in Japanese Black cattle. Mamm Genome 2004; 14:142-148. [ Links ]
Zhang S, Knight T, Reecy J, Beitz D. DNA polymorphism in bovine fatty acid synthase are associated with beef fatty acid composition. Anim Genet 2008; 39:62-72. [ Links ]