Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Colombiana de Ciencias Pecuarias
Print version ISSN 0120-0690
Rev Colom Cienc Pecua vol.27 no.1 Medellín Jan./Mar. 2014
ORIGINAL ARTICLES
Association of BoLA-DRB3 and TLR4 alleles with subclinical mastitis in cattle from Colombia¤
Asociación de los alelos BoLA-DRB3 y TLR4 con mastitis subclínica en vacas de Colombia
Associação de Bola-DRB3 alelos e TLR4 com mastite subclínica em vacas da Colômbia
Nicolás F Ramírez1*, DVM, MSc, (c) Dr. Sc. An.; Alba Montoya2, Biol, MSc; Mario F Cerón-Muñoz2, Zoot, PhD; David Villar3, DVM, PhD; Luis G Palacio1, DVM, PhD.
1Línea de epidemiología y salud pública veterinaria, Grupo de Investigación Centauro, Escuela de Medicina Veterinaria, Facultad de Ciencias Agrarias, Universidad de Antioquia, Calle 70 No. 52-21, Medellín, Colombia.
2Grupo de Investigación GaMMa, Facultad de Ciencias Agrarias, Universidad de Antioquia, Calle 70 No. 52-21, Medellín, Colombia.
3Grupo de Investigación Biogenesis, Facultad de Ciencias Agrarias, Universidad de Antioquia, Calle 70 No. 52-21, Medellín, Colombia.
* Corresponding author: Nicolas F Ramirez. Escuela de Medicina Veterinaria, Facultad de Ciencias Agrarias, Universidad de Antioquia. Tel: (054) 2199131. Email: nramirez@agronica.udea.edu.co
(Received: February 13, 2013; accepted: October 21, 2013)
Summary
Background: molecular markers for genetic resistance can be used to control mastitis in dairy cattle. The Major Histocompatibility Complex and the Toll-like receptor 4 (TLR4) are two promising genes that warrant investigation. Objective: to identify associations between genotypes of BoLA-DRB3 locus and T4CRBR2 fragment and subclinical mastitis (SM). Methods: 996 lactating cows from 32 herds comprising Holstein (80%), Holstein x Jersey cross (12.5%), and other crosses (7.5%) were evaluated monthly during two years, diagnosed for SM and genotyped for the second exon of BoLA DRB3 and the TLR4 coreceptor-binding region 2 (T4CRBR2) using a Polymerase chain reaction-restriction fragment length polymorphism technique (PCRRFLP). The association between candidate alleles and subclinical mastitis was measured by logistic regression. Results: the most frequently observed alleles for BoLA-DRB3 were DRB3.2 *8, *22, *24, *16, *10, *23, *gba, *11, *2, *mbb, *jba, *3, and *15, accounting for 58.9% of the population. Frequencies for T4CRBR2 alleles A and B were 0.352 and 0.647, respectively. Based on 57,408 observations during the period, the mean SM prevalence was 16.2% (95% CI 13.0 and 19.4) per udder quarter and 37.6% (95% CI 32.1 and 43.2) per cow. The predominant microorganisms isolated from SM quarters were Streptococcus agalactiae and Coagulase-Negative Staphylococci (CNS). Allele DRB3.2 *23 was associated with SM occurrence and CNS infection. No alleles were associated with Streptococcus agalactiae infection. Allele *mbb was associated with occurrence of CNS infection and alleles *jba and *15 were associated with resistance to CNS infection. No significant relationship between T4CRBR2 and SM was observed. Conclusion: DRB3.2 gen may play an important role in the occurrence of SM and certain alleles may confer resistance to specific pathogens.
Key words: genetic markers, mammary gland, marker-assisted selection.
Resumen
Antecedentes: los marcadores moleculares genéticos de resistencia para mastitis bovina son una herramienta para el control de la enfermedad en rebaños lecheros. Los genes del Complejo Mayor de Histocompatibilidad y el Receptor tipo Toll 4 (TLR4) son dos genes candidatos promisorios que justifica investigar. Objetivo: identificar asociaciones entre los genotipos del locus BoLA-DRB3 y del fragmento T4CRBR2 con la ocurrencia de mastitis subclínica. Métodos: 996 vacas lactantes de 32 hatos de las razas Holstein (80%), Holstein x Jersey (12,5%) y otros cruces (7,5%), fueron visitadas mensualmente por dos años, diagnosticadas para mastitis subclínica y genotipificadas para el segundo exón del DRB3 y para la región 2 de unión al correceptor del TLR4 (T4CRBR2) por medio de las técnicas de Reacción en cadena de la polimerasa y de Longitud del polimorfismo del fragmento de restricción (PCR-RFLP). La asociación entre los alelos candidatos y la mastitis subclínica se midió por regresión logística. Resultados: los alelos más frecuentes para el DRB3.2 fueron *8, *22, *24, *16, *10, *23, *gba, *11, *2, *mbb, *jba, *3 y *15, que suman el 58,9% del total en la población. Las frecuencias para los alelos A y B del T4CRBR2 fueron de 0,352 y 0,647, respectivamente. Basados en 57.408 observaciones, la prevalencia de MS a nivel de cuarto fue 16,2% (95% IC 13,0 y 19,4) y a nivel de vaca fue de 37,6% (95% IC 32,1 y 43,2). Los microorganismos más frecuentes fueron Streptococcus agalactiae y Estafilococo Coagulasa Negativo (ECN). El alelo DRB3.2 *23 fue el más asociado con la ocurrencia de MS y con la infección por ECN. No se hallaron alelos asociados a infección con mastitis por Streptococcus agalactiae. Con respecto a la infección por ECN, el *mbb se asoció con la ocurrencia y los alelos *jba y *15 se asociaron con resistencia. No se observó asociación entre T4CRBR2 y MS. Conclusión: el gen DRB3.2 puede jugar un papel importante en la presencia de MS y ciertos alelos pueden conferir resistencia a patógenos específicos.
Palabras clave: glándula mamaria, marcadores genéticos, selección asistida por marcadores.
Resumo
Antecedentes: o uso de marcadores moleculares de resistência para mastites permite controlar esta doença em rebanhos leiteiros. Os genes Toll Like Receptor 4 (TLR4) e o Complexo Mayor de Histocompatibilidade são dois genes candidatos promissórios que justifica pesquisar. Objetivo: identificar associações entre genótipos do locus BoLA-DRB3 e do fragmento T4CRBR2 com a ocorrência de mastite subclínica. Método: 996 vacas em lactação de 32 rebanhos da raça Holandesa (80%), Holandesa x Jersey (12,5%) e outras cruzas (7,5%) foram visitadas mensalmente por dois anos, diagnosticadas para mastites subclínica e genotipadas para o exon segunda BoLA DRB3 e região 2 da ligação co-receptor TLR4 (T4CRBR2) através das técnicas de Reação em Cadeia da Polimerase e do Polimorfismo de Comprimento do Fragmento de Restrição (PCRRFLP). A associação entre alelos candidatos e mastite subclínica foi realizada por meio de regressão logística. Resultados: os alelos mais frequentes *8, *22, *24, *16, *10, *23, *gba, *11, *2, *mbb, *jba, *3 e *15, com um 58,9% do total da população. As frequências dos alelos A e B do T4CRBR2 foram 0,352 e 0,647, respectivamente. Com base em 57.408 observações, a prevalência da SM em quartos mamários foi de 16,2% ((IC 95% 13,0 e 19,4) e ao nível de vaca foi de 37,6% (IC 95% 32,1 e 43,2). Os microrganismos mais comuns foram: Streptococcus agalactiae e Estafilococos Coagulase-negativo, ECN. O alelo DRB3.2 *23 foi o mais associado com a ocorrência de SM e com a infecção por ECN. Não foram encontrados alelos associados à infecção por Streptococcus agalactiae. Em relação à infecção por ECN, o *mbb esteve associado com ocorrência e os alelos *jba e *15 estiveram associados com resistência. Não existiu associação entre MS e os alelos do T4CRBR2. Conclusão: o gene DRB3.2 bovino pode desempenhar um papel importante na presencia de MS e alguns alelos podem conferir resistência à patógenos específicos.
Palavras chave: glêndula mamaria, marcadores genéticos, seleção assistida por marcadores.
Introduction
Despite efforts to control and prevent mastitis through udder health programs, mastitis remains the dairy industry's costliest disease (Francoz et al., 2012). Reducing mastitis losses is necessary to keep milk producers economically competitive and maintain costs of dairy products within reasonable limits. Current methods to reduce mastitis incidence, such as therapeutic and prophylactic measures, are not totally effective. Another approach could be to improve cows' natural geneticresistance to udder pathogens (Detilleux, 2002).
Interest in genetic markers for disease resistance has increased, particularly in the Major Histocompatility Complex (MHC) genes, also known as bovine leukocyte antigen (BoLA), which has been widely studied. This complex has been mapped to chromosome 23 and consists of classes I, II and III, which span about 2.5 Mbp (Andersson and Davies, 1994). Most studies associating the occurrence of infectious diseases in cattle with gene candidates have focused on gene DRB3 of the BoLA IIa region which encodes for the antigen binding groove of the molecule and is highly polymorphic (Xu et al., 1993; Dietz et al., 1997a; Dietz et al., 1997b; Duangjinda et al., 2009). Although breeding programs would benefit from selecting for alleles associated with resistance (or host defense mechanisms) to multiple pathogens, conflicting results have been reported on whether specific alleles offer protection or susceptibility to mastitis. Consequently, further studies are warranted to clarify the role of DRB3 alleles in SM.
Other candidate genes with key roles in immune cells are the Toll-like receptor (TLR) family, which mediate recognition of numerous microbial components (Taro and Akira, 2005). Multiple TLRs detect several features of a microbe simultaneously and in particular, TLR4 has been characterized for detecting the structure of the Gram-negative bacterial lipopolysaccharide (Underhill and Ozinsky, 2002). The TLR4 gene encodes for a protein of 841 amino acids, of which two domains are for the putative co-receptor-binding regions 1 and 2 (T4CRBR1 and T4CRBR2). The genetic polymorphism of these regions has been characterized by PCR-Single-strand conformation polymorphism and PCR-RFLP (Wang et al., 2007). Interestingly, mastitis in cows strongly increases mRNA expression of TRL4 (Goldammer et al., 2004), but whether that improves pathogen perception and signaling still remains to be elucidated.
The objective of this study was to identify associations between the most frequent genotypes of BoLA-DRB3 locus and fragment T4CRBR2 in cows with the occurrence of subclinical mastitis.
Materials and methods
Ethical considerations
This study was approved by the Ethics Committee for Animal Experimentation of the University of Antioquia (Act number 48, December 12, 2008).
Herd selection
A convenience sample of 32 herds was selected from the 3,049 registered dairy farms from the six municipalities that comprise the specialized dairy production area in the high plains of northern Antioquia, Colombia (Entrerrios, Belmira, Santa Rosa de Osos, San Jose de la Montaña, San Pedro de los Milagros, and Donmatias). Within each municipality, herds were selected based on the following typical management conditions in the region, including herd size, type of milking system, nutrition program, and breed. Other aspects taken in to account when selecting the herds were easy road access, on-farm cooling tanks, proper cow identification, and owner's willingness to collaborate with sample collection. All cows in each selected herd were included in the study.
Visit protocol and sample collection
Farms were visited each month for 2 years (January 2009 to December 2010) for collection of milk samples during the evening milking. A California Mastitis Test (CMT) was performed on all milking cows that were more than 4 days postpartum. Udder and teats were prepared by the farmer using the normal routine. Subsequently, the first streams of milk were discarded before collection. CMT was performed as instructed by the manufacturer. When a positive or suspect CMT resulted, the teat ends were disinfected with cotton swabs drenched in 70% alcohol, and duplicate samples (approximately 5 ml each) were collected from every quarter and later submitted for a somatic cell count (SCC) using a DeLaval Cell Counter (DCC) (DeLaval Stockholm, Sweden). Any sample containing SCCâ¥200,000 cells/mL was submitted for bacterial culture. The SCC test and bacterial cultures were performed at the Microbiology Laboratory of the Veterinary Medicine School of the University of Antioquia.
Bacteriological culture
Samples were analyzed using standard laboratory methods for microbiological analysis (MacFaddin, 2000; Winn et al., 2006). Growth of three or more bacterial species was considered contaminated and discarded from the analysis (Parker et al., 2008). Performed tests were intended to identify the following microorganisms: Staphylococcus aureus, Coagulase-negative Staphylococci (CNS), Streptococcus agalactiae, Streptococcus uberis, Streptococcus dysgalactiae, Streptococcus pyogenes, Corynebacterium spp., Escherichia coli, Enterobacter aerogenes, Klebsiella spp., Pseudomonas spp., and yeast (Candida spp.).
Case definition
An udder quarter was considered affected by SM if it had a trace CMT score or higher and a subsequent SCC 2 ≥ 00,000 cells/mL. A cow was classified as having SM if one or more quarters were affected by SM.
For pathogen prevalence, a quarter was considered infected if it was diagnosed as having SM and either one or two pathogens were isolated from the milk sample. Quarters with negative CMT score were not sampled and hence were considered as non-infected in the analysis. A cow was considered infected if one or more quarters were infected. Only Streptococcus agalactiae and CNS infections were analyzed in detail.
Blood collection and DNA extraction
EDTA vacutainer tubes were filled with 10 mL blood collected from the tail veins. Genomic DNA was isolated from white blood cells using the salting-out method as previously described (Miller et al., 1988).
Amplification of gene DRB3.2
Amplification of gen RB3.2 (GenBank accession No. 282530) was performed by Nested polymerase chain reaction (PCR) with two sequential reactions, the first round using primers HLO30 (5'-ATC CTC TCT CTG CAG CAC ATT TCC-3') and HL031 (5'TTT AAA TTC GCG CTC ACC TCG CCG CT 3'), followed by a second reaction using primers HLO30 (5'-ATC CTC TCT CTG CAG CAC ATT TCC-3') and HLO32 (5'-TCG CCG CTG CAC AGT GAA ACT CTC-3'), as suggested by Gilliespie et al. (1999).
The first reaction was carried in 10 μL total volume containing 50 ng genomic DNA, 1X PCR buffer (10 mM Tris-HCL pH 9.0; 50 mM KCl; 0.1% Triton® X-100), MgCl2 (2.5 mM), dNTPs (0.2 mM), 0.5 mM of each primer and 1.5 units of Taq DNA polymerase (Fermentas®, Thermo Scientific, Waltham, MA, USA). For the second reaction, the composition in a total of 40 μL was: 4X buffer (10 mM Tris-HCL pH 9.0; 50 mM KCl; 0.1% Triton® X-100), MgCl2 (1.75 mM), dNTPs (0.25 mM), 0.5 mM for each primer and 2.0 units of Taq DNA Polymerase (Fermentas®, Thermo Scientific, Waltham, MA, USA), and 2 μl of the first reaction product. The reactions were performed in a thermal cycler (Bio-Rad C-1000®, Bio-Rad Laboratories, Hercules, California, USA) using the following cycling profile: initial denaturation at 94 °C for 5 min; 35 cycles at 94 °C for 45 s, 60 °C for 1 min, 72 °C for 1 min; and a final extension at 72 °C for 7 min. The final product was a 284 bp segment that was digested separately with 3 restriction endonucleases: RsaI, HaeIII and BstyI, as described by Van Eijk et al. (1992). Digestions were performed as prescribed by the manufacturer (FastDigest®, Fermentas®, Thermo Scientific, Waltham, MA, USA). The resulting fragments were resolved in a 8% denaturating polyacrylamide gel by running at 1600/1900 V, 25/60 mA for 2.5 hours and then stained with nitric acid as described by Budowle et al., (1991). The designation into different alleles was made using a reference ladder (Low Range DNA Ladder, O'gene rulerTM, Fermentas®, Thermo Scientific, Waltham, MA, USA) according to the allelic nomenclature described by Van Eijk et al. (1992).
Amplification of gene TLR4 (GenBank accession No. DQ839566)
The coreceptor-binding region 2 of the gen was amplified using the following primers: forward 5'-AGACAGCATTTCACTCCCTC-3' and reverse -5'-ACCACCGACACACTGATGAT-3'. The PCR reactions were carried out in 20 μL total volume containing 50 ng genomic DNA, 1X buffer (10 mM Tris-HCL pH 9.0; 50 mM KCl; 0.1% Triton® X-100), MgCl2 (2.0 mM), dNTPs (0.2 mM), each of the primers (0.25 μM) and 0.5 U Taq DNA Polimerase (Fermentas®, Thermo Scientific, Waltham, MA, USA). The reactions were conducted in a thermal cycler (Bio-Rad C-1000®, Bio-Rad Laboratories, Hercules, California, USA) using the following protocol: initial denaturation at 94 °C for 5 min, 35 cycles at 94 °C for 30 s, 61.5 °C for 30 s, 72° C for 50 s; and a final extension at 72 °C for 6 min. The final product was a 382 pb amplicon digested with AluI FastDigest® enzyme as prescribed by the manufacturer (Fermentas®, Thermo Scientific, Waltham, MA, USA). The sample was then electrophoresed in agarose gel and DNA band patterns were observed under a transiluminator (UV Transilluminator UVP) and genotypes identified based on an allelic ladder (Low Range DNA Ladder, O'gene rulerTM, Fermentas®, Thermo Scientific, Waltham, MA, USA).
Statistical analysis
Analysis of allele frequencies. The frequencies of each restriction product from RsaI, HaeIII y BstYI (PsuI) enzymes were calculated using PopGen32 (Yeh et al., 2000). Due to the highly polymorphic nature of BoLAâDRB3.2, only alleles with >2% frequency were used for the analysis and considered as independent variables. The dependent variables were the disease state: SM, infection by Streptococcus agalactiae, and CNS infection. The polymorphism for the T4CRBR2 region of TLR4 gene was assessed by calculating the frequencies for the A and B alleles.
Logistic regression analysis. Data was examined for biologically implausible entries and any erroneous entries were removed. Descriptive statistics were computed for all variables of interest using standard methods. Observations were stratified by municipality and sampling weights were computed as the inverse of the probability of the herd being selected in the municipality (i.e., # of herds in municipality divided by # of herds selected in municipality). The clustering and repeated measures structure of the collected data (multiple observations from quarters clustered within cows, clustered within herds) were taken into account by using variance linearization estimation procedures with herds identified as the primary clustering unit (this is equivalent to using robust standard errors which are allowed to vary across clusters of herds) using the survey package of StataTM (StataCorp 2011, College Station, TX, USA). The model response variables were SM, Streptococcus agalactiae, and CNS infection (presence or abscense). The effect of cluster was at random and each one of the risk factors (breed, month in lactation, parity, and the DRB3.2 and TLR4 alleles) were fixed effects.
The survey logistic model took the general form:
(Y) ~ binary outcome (probability π),
Logit (π) = intercept + β1b + β2-14DRB3.2 +
β15TLR4 + β16 lm + β17 p + e
Where Y is the outcome variable; π is the fitted probability of the outcome; β1 to β17 are the coefficients associated with each covariate: b is breed covariate, DRB3.2 is DRB3.2 alleles (*8, *24, *22, *15, *3, *jba, *mbb, *11, *2, *gba, *23, *10, *16) covariate, TLR4 is the covariate TLR4 allele (*T4CRBR2), p is parity covariate, lm is lactation month covariate, and e is the random residual effect.
Unconditional logistic analyses between each risk factor and the outcomes were computed, and associations with p<0.15 were retained for consideration in multivariable models. Multivariable logistic regression models were constructed with variables remaining in the model if they had a Wald test p<0.05. The potential confounding effect of breed was evaluated by refitting the final models with breed omitted to see if the coefficients for other predictors changed substantially. Results from the final models are presented as odds ratios (ORs) along with their 95% confidence intervals. If OR was greater than 1, the factor was considered as a risk factor for causation of the disease. When OR was less than 1, the factor should be viewed as a sparing or a resistant factor (Martin et al., 1987). Model adequacy was assessed by the Hosmer-Lemeshow goodness of fit test (Hosmer and Lemeshow, 2000) for regression analysis (Vittinghoff et al., 2012). It should be highlighted that the Hosmer Lemeshow goodness of fit test was performed for all regression models, observing a proper adjustment for each. Analysis was done using the Stata's survey command of Stata® (StataCorp 2011, College Station, TX, USA).
Results
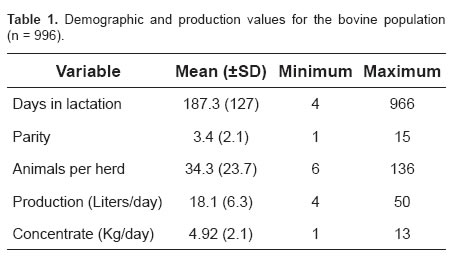
A total of 57,408 observations were made at the quarter level, accounting for 996 cows in 32 herds, 80.0% of which were Holsteins, 12.5% Holstein x Jersey, and 7.5% other crossbreds. The mean (±SD) of the population demographic and production parameters is shown in table 1. Mastitis prevalence at the quarter and cow levels were 16.2% (95% CI 13.0 and 19.4) and 37.6% (95% CI, 32.1 and 43.2), respectively. The most frequently isolated bacteria were Streptococcus agalactiae and CNS in 32.4% and 17.3% of the cases, respectively (data not shown).
Allele frequencies of BoLA DRB3.2 genes digested with RsaI, HaeIII and BstYI
The PCR product of the DRB3.2 locus was a 284 bp amplicon. Digestion with RsaI resulted in 92 genotypes and 19 alleles identified as a-o, r, s, u, and w, with frequencies ranging from 0.0005 for allele r, to 0.1898 for allele f. The effective allele number was 7.9 and the heterozygosity value was 0.8243.
HaeIII enzyme resulted in 26 genotypes and 8 alleles identified as a-f, h and i. Frequencies varied from very low for allele c (0.0040) to high for allele a (0.5371), with 2.74 effective number of alleles and 0.6345 heterozygocity value.
The BstY (PsuI) restriction enzyme yielded 8 genotypes with 4 of the 5 alleles (a, b, d and e). The lowest frequent was allele d (0.0040) and the most frequent was allele b (0.6662). The effective number of alleles was 1.95 and the heterozygosity value was 0.4679.
Allele frequencies of TLR4 (T4CRBR2 region)
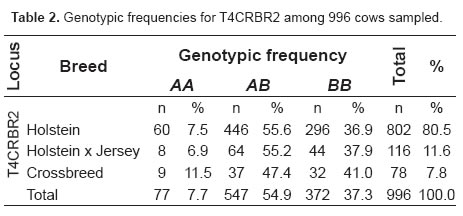
By digestion with AluI enzyme, a segment of 382 bp was amplified from locus T4CRBR2 by PCR. Two alleles (*A y *B) were obtained with frequencies of 0.3522 and 0.6478, respectively. The effective number of alleles was 1.83 and heterozygosity was 0.5515. Genotypic frequencies of the T4CRBR2 locus for each breed examined are shown in table 2.
Allele frequencies of BoLA DRB3.2
A total of 149 alleles were found from 996 cows sampled; those with frequencies below 0.02 are not reported. When alleles with frequencies below 2% were removed from the analysis of association with SM, only 381 cows remained in the regression analysis. The distribution of those alleles with frequencies > 2% in 996 cows ranged from 0.048 to 0.123 (Table 3). In order of decreasing frequencies, the most common alleles were *8 (0.123), *22 (0.095), and *24 (0.070).
Logistic regression analysis
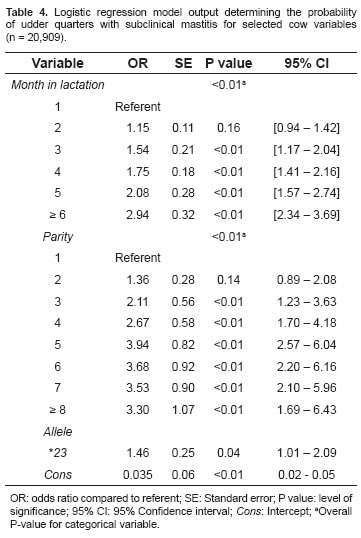
An initial logistic regression analysis of DRB3.2 and TLR4 (T4CRBR2) alleles and some factors that included data from all 381 cows (without the breed variable) showed no changes in OR or standard errors for genetic variables. No association between SM and breed in the final model was observed (p>0.05). Regarding months in lactation, a significant (p<0.01) linear association occurred between time in lactation and SM risk, which was significant in the third month and beyond. Similarly, parity was significantly (p<0.01) associated with increased risk of suffering SM from third parity and beyond (p<0.01). Regarding T4CRBR2 alleles, no association was found with SM, St. agalactiae, or CNS infection. For the DRB3.2 alleles, only allele *23 behaved as a risk factor for SM and showed significant (p<0.05) OR of 1.46 (Table 4).
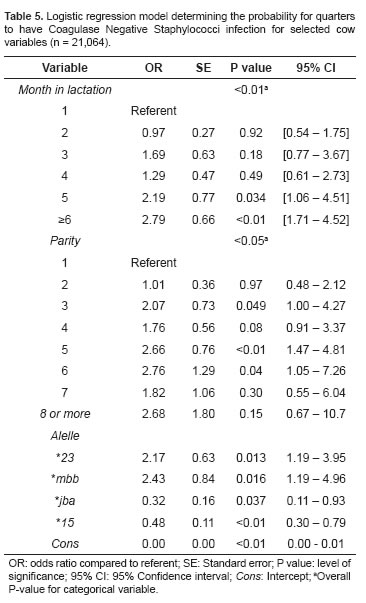
No association was found between DRB3.2 alleles and Streptococcus agalactiae infection in the mammary gland. The analysis of DRB3.2 alleles and some cow factors associated to the infection by CNS in the mammary gland are shown in table 5. Breed was not a significant risk factor for the occurrence of such infection (p>0.05). A significantly high infection risk was observed for parities 3 (p = 0.049), 5 (p<0.01), and 6 (p = 0.04). Months of lactation, when referenced with time between birth and first month of lactation, presented an increased risk of CNS infection at 5 or more months (p<0.05). For CNS infection the DRB3.2 genes, alleles *jba and *15 behaved as protective factors with 0.32 (p = 0.04) and 0.48 (p<0.01) OR, respectively, and alleles *23 and *mbb behaved as a risk factor with 2.17 (p = 0.013) and 2.43 (p = 0.016) OR, respectively.
Discussion
This study analyzed the potential association of TLR4 (T4CRBR2) gene and DRB3.2 alleles with the occurrence of SM, Streptococcus agalactiae, and CNS related infections. In terms of allelic frequencies, the results of digestion with 3 restriction endonucleases showed that DRB3.2 locus was highly polymorphic. Results of digestion with RsaI endonuclease coincided with those previously assigned to letters a-o, r, s, (Van Eijk et al., 1992), u (Gelhaus et al., 1995) and w (Maillard et al., 1999). Digestion with HaeIII enzyme coincided with pattern a to f described by Van Eijk et al. (1992), h (Gelhaus et al., 1995), and i (Roslin, 2002). Similar to the fragments attained with RsaI, HaeIII enzyme showed high heterozygosity value, implying a high degree of genetic variation in this locus. The alleles resulting from digestion with BstY (PsuI) endonuclease coincided with those reported by Van Eijk et al. (1992).
This study found 149 distinct DRB3.2 alleles, though frequencies below 2% were considered irrelevant for the analysis of association with SM, S. agalactiae and CNS infection, and therefore excluded to avoid confounding results. Demonstration of the existence of new DRB3.2 alleles should be accompanied by proper sequencing, which was beyond the scope of this study.
The number of alleles was high, but according to Lewin and Van Eijk (1994), when all restriction fragment patterns for each enzyme (RsaI, BstY1, HaeIII) are considered, a total of 2048 combination of sites are possible, and twice that number of alleles is possible if a 3 base pair deletion is included as a source of variation (Lewin and Van Eijk, 1994). The high polymorphism of DRB3.2 found was probably accentuated by the large number of animals in the sample (n = 996) and also by the different breeds and crosses represented. The sample was mainly composed by Holstein and Holstein x Jersey crossbreds; the category ''other breeds or crosses'', despite being the group with the lowest number of animals, was represented by a great diversity that included Ayrshire, Swedish Red, Swiss Brown, Jersey, and several crosses of Holstein with other breeds such as Ayrshire, Angus, Blanco Orejinegro (BON), Brahman and Gir.
High polymorphisms in DRB3.2 from cattle in other Colombian studies were reported by Zambrano et al. (2011), who identified 23 and 18 alleles in 66 pure Holstein and 25 BON cows, respectively. Martinez et al. (2005) found 35 and 11 alleles in 140 BON and 22 Brahman cows, respectively. Hernandez, (2010) reported 41 alleles in 360 cows from different breeds, mostly creole Colombian cattle; Giovambattista et al. (2013) reported 24 (22 reported and two new) in Hartón del Valle breed. In Mexico, high polymorphism was also reported by Fernandez et al. (2008) who identified 52 DRB3.2 alleles in 98 cattle, and in Thailand Duangjinda et al. (2009) reported 40 different alleles in 409 Holstein x Zebu cows. In particular, the high frequency observed for allele *23 was also reported by Zambrano et al. (2011) in Holstein and BON x Holstein cows from Colombia. In this study, the high frequencies of alleles DRB3.2 *8, *22 and *24 in Holstein cows was similar to that reported by other authors (Dietz et al., 1997a; Sharif et al., 1998; Rupp et al., 2007) and by Starkenburg et al. (1997) who reported that alleles *8, *22,*23, *24 and *27 accounted for 66.8% of all alleles. Van Eijk et al. (1992) and Gelhaus et al. (1995) reported high frequencies for alleles *8 and *11 in Holstein cows. With regards to T4CRBR2, the predominant allele was B in the three cattle populations, genotypic frequency for AB was the highest in both loci, with AA being the lowest, in agreement with the findings by Wang et al. (2007).
Relationship between bovine subclinical mastitis, S. agalactiae and CNS infection and the polymorphisms of TLR4 (T4CRBR2) and DRB3.2 genes
In the logistic regression analysis, breed was not associated with SM, S. agalactiae or CNS infection. An increased risk for SM was observed as the number of months in lactation and parities increased. Number of births and stage of lactation have also been previously associated with SM, and is likely attributed to the normal tissue deterioration from manipulation and aging (Breen et al., 2009). No association was observed between genotypes of T4CRBR2 and SM, Streptococcus agalactiae, or CNS infection in the logistic regression model. This results agree with those of Wang et al. (2007), who found no significant association between genotypes of locus T4CRBR2 and Somatic Cell Score, as an index for mastitis. No literature reports could be found on possible associations between T4CRBR2 and Streptococcus agalactiae or CNS infection. However, the identification of TLR-4 on milk fat globule membranes suggest a direct role for the mammary gland parenchyma in pathogen detection (Reinhardt and Lippolis, 2006). It has also been proposed that TLR4 is likely involved in the signal transduction pathway that mediates the pathogenesis of E. coli mastitis (De Schepper et al., 2008).
In this study, allele *23 was linked to SM susceptibility, which concurs with other findings that report the association of this allele to increased prevalence of SM caused by Streptococcus dysgalactiae (Hameed et al., 2008), higher SCC in cows in their third or higher parity (Dietz et al., 1997a) and occurrence of severe mastitis (Sharif et al., 1998). By contrast, Baltian et al. (2012) found this allele to have a protective effect against mastitis diagnosed by SCC. The diverging results in the literature linking different DRB3.2 alleles to resistance or susceptibility towards SM may be attributed to multiple reasons such as: differences in pathogens, genetic background, environmental factors, interactions between the former variables, and/or criteria established to diagnose mastitis (Sharif et al., 1998).
When regression analysis was restricted to cows infected with CNS, alleles *jba and *15 were linked to resistance from CNS; yet, no literature reports linking these alleles to either pathogen could be found. However, Duangjinda et al. (2009) reported allele *15 is associated to mastitis resistance in Holstein x Zebu dairy cows. Alleles *23 and *mbb were associated with susceptibility to CNS infection in the mammary gland. There is molecular evidence regarding the potential association between allele *23 and the occurrence of CNS-induced clinical mastitis. A significant association between the presence of glutamic acid at position β74 and occurrence of mastitis caused by Staphylococcus spp. with a relative risk of 11 was detected. (Sharif et al., 2000). No literature reports could be found linking the *mbb allele to any mastitis causing pathogen.
We found an association between allele DRB3.2 *23 and the risk of suffering SM. Alleles DRB3.2 *23 and *mbb were related to susceptibility to infection caused by CNS, and *jba and *15 were related to resistance. DRB3.2 gen may play an important role in the occurrence of SM and certain alleles may confer resistance to specific pathogens.
¤ To cite this article: Ramírez NF, Montoya A, Cerón-Muñoz MF, Villar D, Palacio LG. Association of BoLA-DRB3 and TLR4 alleles with subclinical mastitis in cattle from Colombia. Rev Colomb Cienc Pecu 2014; 27:18-28.
Acknowledgements
The authors wish to express their gratitude to the Ministerio de Agricultura y Desarrollo Rural de Colombia, Universidad de Antioquia, Cooperativa Lechera de Antioquia, Colanta, and Federación de Asociaciones de Ganaderos de Antioquia (FAGA) for financing the present study. The authors also thank the Sustainability Project 2013-2014 (Estrategia de sostenibilidad CODI 2013-2014, University of Antioquia).
References
Andersson L, Davies CJ. The major histocompatibility complex. In: Goddeeris BML, Morrison WI, editors. Cellmediated immunity in ruminants. Boca Raton: CRC Press, Inc.; 1994. pp. 37-57. [ Links ]
Baltian LR, Ripoli MV, Sanfilippo S, Takeshima SN, Aida Y, Giovambattista G. Association between BoLA-DRB3 and somatic cell count in Holstein cattle from Argentina. Mol biol rep 2012; 39:7215-7220. [ Links ]
Breen JE, Green MJ, Bradley AJ. Quarter and cow risk factors associated with the ocurrence of clinical mastitis in dairy cows in the United Kingdom. J Dairy Sci 2009; 92:2551-61. [ Links ]
Budowle B, Chakraborty R, Giusti AM, Eisenberg AJ, Allen RC. Analysis of the VNTR locus D1S80 by the PCR followed by high-resolution PAGE. Am J Hum Genet 1991; 48:137-144. [ Links ]
De Schepper S, De Ketelaere A, Bannerman DD, Paape MJ, Peelman L and Burvenich C. The toll-like receptor-4 (TLR-4) pathway and its possible role in the pathogenesis of Escherichia coli mastitis in dairy cattle. Vet Res 2008; 39:2-23. [ Links ]
Detilleux JC. Genetic factors affecting susceptibility of dairy cows to udder pathogens. Vet Immunol Immunopathol 2002; 88:103-110. [ Links ]
Dietz AB, Cohen ND, Timms L, Kehrli ME. Bovine lymphocyte antigen class II alleles as risk factors for high somatic cell counts in milk of lactating dairy cows. J Dairy Sci 1997a; 80:406-412. [ Links ]
Dietz AB, Detilleux JC, Freeman AE, Kelley DH, Stabel JR, Kehrli ME. Genetic association of bovine lymphocyte antigen DRB3 alleles with immunological traits of Holstein cattle. J Dairy Sci 1997b; 80:400-405. [ Links ]
Duangjinda M, Buayai D, Pattarajinda V, Phasuk Y, Katawatin S, Vongpralub T, Chaiyotvittayakul A. Detection of bovine leukocyte antigen DRB3 alleles as candidate markers for clinical mastitis resistance in Holstein x Zebu. J Anim Sci 2009; 87:469-476. [ Links ]
Fernandez I, Rios J, Gayosso V. Polymorphism of locus DRB3.2 in populations of Creole Cattle from Northern Mexico. Genet Mol BioL 2008; 31:880-886. [ Links ]
Francoz D, Bergeron L, Nadeau M, Beauchamp G. Prevalence of contagious mastitis pathogens in bulk tank milk in Quebec. Can Vet J 2012; 53:1071-1078. [ Links ]
Gelhaus A, Schnittger L, Mehlitz D, R Horstmann D, C Meyer G. Sequence and PCR-RFLP analysis of 14 novel BoLA-DRB3 alleles. Anim Genet 1995; 26:147-153. [ Links ]
Gilliespie BE, Jayarao BM, Dowlen HH, Oliver SP. Analysis and frequency of bovine lymphocyte antigen DRB3.2 alleles in Jersey cows. J Dairy Sci 1999; 82:2049-2053. [ Links ]
Giovambattista G, Takeshima SN, Ripoli MV, Matsumoto Y, Franco LA, Saito H, Onuma M, Aida Y. Characterization of bovine MHC DRB3 diversity in Latin American Creole cattle breeds. Gene 2013; 519:150-158. [ Links ]
Goldammer T, Zerbe H, Molenaar A, Schuberth HJ, Brunner RM, Kata SR, Seyfert HM. Mastitis increases mammary mRNA abundance of beta-defensin 5, toll-like-receptor 2 (TLR2), and TLR4 but not TLR9 in cattle. Clin Diagn Lab Immunol 2004; 11:174-185. [ Links ]
Hameed K, Sender G, Korwin-Kossakowska A. An association of BoLA alleles DRB3.2*16 and DRB3.2*23 with occurrence of mastitis caused by different bacterial species in two herds of dairy cows. Anim Sci Pap Rep 2008; 26:37-48. [ Links ]
Hernandez D. Asociación del locus BoLA-DRB3.2 con el virus de la leucosis Bovina en razas criollas y colombianas. Facultad de Ciencias Agropecuarias. Universidad Nacional de Colombia, Palmira. 2010; [access date: June 15, 2011] URL: http://www.bdigital.unal.edu.co/2137/1/740803.2010.pdf [ Links ]
Hosmer DW, Lemeshow S. Applied logistic Regression. John Wiley & Sons: New York; 2000. [ Links ]
Lewin H and M Van Eijk, inventors. Methods for testing bovine for resistance or susceptibility to persistent lymphocytosis by detecting polymorphism in BoLA-DR3 exon 2. United States Pat. No. 5,582,987. 1994; [access date: August 14, 2013] URL: http://www.google.com/patents/US5582987 [ Links ]
MacFaddin J. Biochemical Tests for Identification of Bacteria. 3rd ed. Baltimore: Williams & Wilkins; 2000. [ Links ]
Maillard JC, Renard C, Chardon P, Chantal I, Bensaid A. Characterization of 18 new BoLA-DRB3 alleles. Anim Genet 1999; 30:200-203. [ Links ]
Martin S W, Meek AH, Willeberg P. Veterinary Epidemiology (Principles and Methods). Ames (IA): Iowa State University Press; 1987. [ Links ]
Martinez R, Toro R, Montoya F, Burbano M, Tobón J, Gallego J, Ariza F. Caracterización del locus BoLA_DRB3.2 en ganado criollo colombiano y asociación con resistencia a enfermedades. Arch Zootec 2005; 54:349-356. [ Links ]
Miller SA, Dykes DD, Poleesky HF. A Simple Salting out procedure for extracting DNA from Human Nucleated Cells. Nucleic. Nucleic Acids Res 1988; 16:1215. [ Links ]
Parker KI, Compton CW, Anniss FM, Heuer C, McDougall S. Quarter-level analysis of subclinical and clinical mastitis in primiparous heifers following the use of a teat sealant or an injectable antibiotic, or both, precalving. J Dairy Sci 2008; 91:169-181. [ Links ]
Reinhardt TA, Lippolis JD. Bovine milk fat globule membrane proteome. J Dairy Res 2006; 73:406-416. [ Links ]
Roslin. DRB3 PCR-RFLP analysis in BoLA Nomenclature, International Society for Animal Genetics 2002. [access date: January 24, 2011] URL: http://www.projects.roslin.ac.uk [ Links ]
Rupp R, Hernandez A, Mallard BA. Association of bovine leukocyte antigen (BoLA) DRB3.2 with immune response, mastitis, and production and type traits in Canadian Holsteins. J Dairy Sci 2007; 90:1029-1038. [ Links ]
Sharif S, Mallard BA, Wilkie BN, Sargeant JM, Scott HM, Dekkers JC, Leslie KE. Associations of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) alleles with ocurrence of disease and milk somatic cell score in Canadian dairy cattle. Anim Genet 1998; 29:185-193. [ Links ]
Sharif S, Mallard B, Sargeant J. Presence de glutamine at position 74 of pocket 4 in the BoLA-DR antigen binding groove is associated with occurrence of clinical mastitis caused by Staphylococcus species. Vet Immunol immunopatol 2000; 76:231-238. [ Links ]
Starkenburg RJ, Hansen LB, Kehrli ME, Chester-Jones H. Frequencies and effects of alternative DRB3.2 alleles of bovine lymphocyte antigen for Holsteins in milk selection and control lines. J Dairy Sci 1997; 80:3411-3419. [ Links ]
StataCorp. Stata: Release 12. Statistical Software. StataCorp LP., 2011. [ Links ]
Taro K, Akira S. Pathogen recognition with Toll-like receptors. Curr Opin Immunol 2005; 17:338â344. [ Links ]
Underhill DM, Ozinsky A. Toll-like receptors: key mediators of microbe detection. Curr Opin Immunol 2002; 14:103-110. [ Links ]
Van Eijk M, Stewart-Haynes J, Lewin H. Extensive polyorphism of the BoLA-DRB3 gene distinguished by PCRRFLP. Anim Genet 1992; 23:483-496. [ Links ]
Vittinghoff E, Glidden D, Shiboski S, McCulloch C. Regression Methods in Biostatistics. 2nd ed. New York: Springer; 2012. [ Links ]
Wang X, Xu S, Gao X, Ren H, Chen J. Genetic polymorphism of TLR4 gene and correlation with mastitis in cattle. J Genet Genomics 2007; 34:406-412. [ Links ]
Winn W, Allen S, Janda W, Koneman E, Procop G, Schrenckenberger P, Woods G. Koneman's Color Atlas and Textbook of Diagnostic Microbiology. 6th ed. Baltimore (USA): Lippincott Williams & Wilkins; 2006. [ Links ]
Xu A, van Eijk MJ, Park C, Lewin HA. Polymorphism in BoLA-DRB3 exon 2 correlates with resistance to persistent lymphocytosis caused by bovine leukemia virus. J Immunol 1993; 151:6977-6985. [ Links ]
Yeh F, Yang R, Boyle T, Ye Z. Microsoft Windows-Based Freeware for Population Genetic Analysis Version 1.32 ed. Molecular Biology and Biotechnology Centre. University of Alberta, Edmonton. 2000. [ Links ]
Zambrano J, Echeverry J, Lopez A. Alleles of the BoLA DRB3.2 gene are associated with mastitis in dairy cows. Rev Colomb Cienc Pecu 2011; 24:145-156. [ Links ]