Introduction
Residual feed intake (RFI) is defined as the difference between actual feed intake and the predicted feed intake based on mean requirements for body weight maintenance and the level of production (Koch et al., 1963). Cattle with a lower residual feed intake consume less food and, therefore, the maintenance requirements are lower, while growth seems to be unaffected (Koch et al., 1963; Crews et al., 2005).
Studies have reported some moderate heritability for this trait. Crews et al. (2005) estimated values from 0.26 to 0.30, and Arthur et al. (2001) and Moore et al. (2007) estimated 0.39 and 0.43, respectively; thus, it is expected that selection for RFI results in a genetic change relatively comparable to those obtained with other moderately heritable traits, such as growth. More excitable (temperamental) cattle may be more easily stressed and less efficient (Lindholm- Perry et al., 2014; Friedrich et al., 2015). Furthermore, temperamental animals usually have less body weight gain, reduced reproductive efficiency, inferior meat quality, and higher disease susceptibility, affecting the economic efficiency in any production system (Friedrich et al., 2015). Negative correlations between daily weight gain and temperament, measured as flight speed, chute score, and others, have been found (Haskell et al., 2014). These results have encouraged interest in including bovine temperament as a strategy to select docile and, therefore, more efficient animals (Lindholm-Perry et al., 2014). Aiming to find genetic markers associated with both traits (temperament and feed efficiency), Lindholm-Perry et al. (2014) found that two markers previously associated with cattle average daily gain/ average daily feed intake (ADG/ADFI, respectively), and frame size were also associated with flight speed. Recently, Garza-Brenner et al. (2017) identified two SNPs, rs109576799 and rs43696138 in the DRD3 and HTR2A genes, respectively, that were significantly associated with exit velocity (EV) and temperament score (TS) in Charolais cows.
To contribute to the discovery of molecular markers that better assist in the selection of animals with higher feed efficiency and docile temperament, the objective of this study was to analyze the effect of two temperament-related SNPs (rs109576799 located in the DRD3 gene and rs43696138 in the HTR2A gene) on feeding performance of Mexican beef cattle.
Material and methods
Description of data and samples sources
The present study did not require ethical clearance from the Animal Care and Use Committee because the data were extracted from existing herd books, and the biological samples and genetic materials were obtained from germplasm banks.
Data and hair samples taken from 136 young Angus (AN), Brangus (BR), and Charolais (CH) bulls, with an average age and body weight of 273 ± 38 d and 272 ± 38 kg, respectively, at the beginning of two (April 2016: AN = 42, BR = 29, CH = 19; and July 2016: AN = 21, BR = 12, CH = 13) centralized feed efficiency performance tests based on residual feed intake (RFI) in northern Mexico (Union Ganadera Regional de Chihuahua, UGRCH) were included.
Feeding and temperament evaluation
Animals were subjected to the same feeding protocol, including a pre-trial adaptation period of 20 d and a feeding trial of 70 d. Animals were weighed on two consecutive days at the beginning and at the end of the trial, and every 14 d during the test. Individual feed intake was measured and recorded using the GrowSafe System (GrowSafe System Ltd., Airdrie, AB, Canada). Calculated traits included residual feed intake (RFI) as described by Koch et al. (1963), daily dry matter feed intake (DMFI), feed to gain ratio (F:G), and average daily gain (ADG).
At the time the animals arrived at the UGRCH facilities, temperament was assessed using pen score (PS), and exit velocity (EV). For PS, three evaluators assigned a score on a 5-point scale, where 1 = unalarmed and unexcited animal that walks slowly away from the evaluator, and 5 = very excited and aggressive toward the technician in a manner that requires evasive action to avoid contact between the technician and the animal, as described by Hammond et al. (1996). Exit velocity was assessed following the stimulus of hair sampling in the chute by measuring the rate of travel over a 1.83-m (6 ft) distance with an infrared sensor (FarmTek Inc., North Wylie, TX, USA). The velocity was calculated as EV = distance (m)/time (s) (Burrow et al. 1997; Hammond et al., 1996).
Genotyping and association
Hair samples collected during temperament testing were used for DNA isolation using the Genelute Mammalian Genomic DNA kit (Cat. G1N350, Sigma- Aldrich Co. LLC, St. Louis, Missouri, USA).
Genotyping of the rs43696138 marker located in the HTR2A gene was performed using allelic discrimination in the ABI PRISM 7500 Sequence Detection System, following the manufacturer’s instructions (Applied Biosystems, Foster City, California 94404, USA). Assay conditions were 2 min at 50°C, 10 min at 95°C, and 40 two-step cycles of 92°C for 15 s and 60°C for 1 min. The SNP assay was set up using SDS software and the allelic discrimination settings (ABI Prism 7500 Real-Time Sequence Detection Software).
To genotype the rs109576799 marker (transversion A/C, located in an intronic region, chromosome 1: 59343757 of the DRD3 gene), a PCR/RFLP assay was designed using the sequence, and SNPID rs109576799 primers (F6799 5’-CTGGAGGCCCGGGGAAGAATCA-3’; R6799 5’-GCCCGCCCACACGCCTACTAC-3’) were used for PCR. The reaction mixes comprised 20-100 ng genomic DNA, 1.5 or 2.5 mM MgCl2, 0.1 µM of each primer, 0.4 mM dNTPs, and 2.5 U Go Taq polymerase (Promega Corporation, Madison, WI, USA). A touchdown method was used with an amplification profile consisting of an initial denaturation step of 95°C for 10 min, five three-step cycles of 45 sec at 95°C and 45 sec at 68°C (the temperature was reduced 2°C each cycle) and 45 sec at 72°C, twenty- five cycles of 45 sec at 95°C and 45 sec at 65°C, and finally, one cycle of 45 sec at 72°C. PCR fragments were separated by electrophoresis in a 1.5% agarose gel, stained with SYBR Gold and visualized by UV irradiation. After PCR, digestion reactions were performed following the manufacturer’s protocol for the Fok1 enzyme, and digestion fragments were analyzed on a 2.5% agarose gel (Sigma-Aldrich).
Allelic and genotypic frequencies were estimated using GENEPOP, web version 4.0.10 (Rousset, 2008). The effects of selected SNP genotypes on analyzed variables were assessed by adjusting a general linear model with the GLM procedure of SAS® (Statistical Analysis System) software, version 9.4 (SAS Institute Inc., Cary, NC, USA) (2013), which included the fixed effects of the number of performance tests and genotype for each studied SNP. In the model:
Y ij = μ + G i + H j + ε ij
Where:
Y: is the dependent variable (EV, PS, ADG, DMFI, RFI, F:G).
μ = is the average.
G: is the effect of the i-th genotype for each individual SNP.
H: is the effect of the j-th performance test (1, 2).
ε ij : is the random error.
Least squares means of genotypes were estimated for the SNPs that demonstrated significant effects (p<0.05), and comparisons of the means were performed with the PDIFF option in the LSMEANS statement of the GLM procedure of SAS.
Results
Phenotypic traits
The mean values for temperament (PS and EV), and for feeding performance traits within each breed are presented in Table 1. According to PS and EV mean values, Brangus was identified as the most (p<0.05) temperamental breed, and Angus was identified as the most docile one.
The mean value for DMFI was significantly lower (p<0.05) for Charolais than for the other two tested breeds. For the F:G ratio, Angus presented the highest (p<0.05) value. No significant differences were observed among breeds for ADG and RFI.
Effect of temperament genetic markers on feed efficiency traits
Table 2 shows the allelic frequencies of the two markers evaluated in the three beef cattle breeds. No significant associations were found between the temperament variables (PS and EV), and the markers evaluated. However, for efficiency parameters, only Brangus showed significant association
Table 1 Temperament and feeding performance of Angus, Brangus and Charolais breeds.
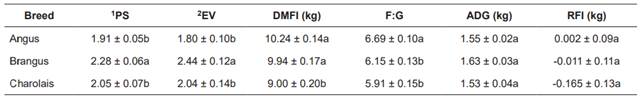
Values are Least square means ± standard error (SE). 1PS: Pen Score, was assigned to animals to describe behavior when approached by a human; scores ranged from 1 (calm) to 5 (most temperamental). 2EV: Exit Velocity, was calculated as velocity = distance (m)/time (s). DMFI: dry matter feed intake; F:G feed to gain ratio; ADG: average daily gain; RFI: residual feed intake. a, b, c: The different letters (a, b) indicate significant differences (p<0.05) between row means.
Table 2 Genotypic and allelic frequencies of rs43696138 and rs109576799 observed in beef cattle breeds.
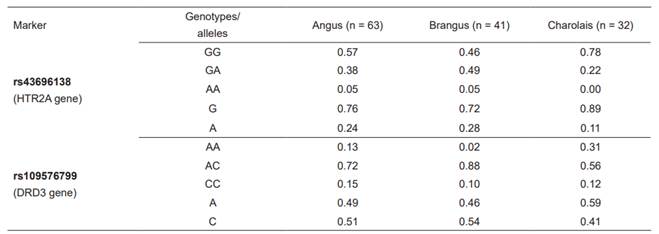
between average daily gain (ADG; p=0.019) and the rs43696138 marker, resulting in higher gains for the homozygous genotype GG (1.69 ± 0.04 kg) compared to the heterozygous genotype GA (1.54 ± 0.04 kg). Due to low allele frequencies of AA genotype among the studied breeds, it was excluded from association analysis.
Discussion
Feed costs represent 50-75% of the total production costs in beef cattle systems. Implementation of feed efficiency for cattle evaluation has been a primary goal in beef cattle systems to reduce feeding costs and negative impacts on the environment (Crews et al., 2005). Both RFI and temperament have been described as complex traits, and some studies have proposed finding the genes that explain and regulate their expression (Crews et al., 2005; Lindholm-Perry et al., 2014).
Phenotypic measures of temperament indicated, as expected, that the Brangus breed had higher values for the temperament variables. Genetic background is an important factor influencing temperament. Bos indicus and Bos indicus crosses tend to be more temperamental than Bos taurus (Burdick et al., 2011). Temperament is most often measured at weaning, and therefore, most of the published literature has focused on the effects of temperament during preweaning and postweaning periods (Burdick et al., 2011). Even when temperament measures were taken at an average age close to weaning of the studied animals, the mean values of the temperament evaluations (i.e., exit velocity) are in a range considered docile in the literature (Burrow et al., 1997; Burdick et al., 2011). Burdick et al. (2011) reported that cattle with an EV of >2.4 m/s are temperamental, while cattle with an EV of <1.9 m/s are calm.
Residual feed intake is a feed efficiency measure that is calculated as the difference between the actual dry matter intake of an animal and its predicted dry matter intake for a given level of maintenance and production (Llonch et al., 2016). Cattle with calm temperaments have been found to have greater average daily gain (ADG) (Burrow et al., 1997); even so, no significant differences were observed for RFI and ADG between breeds.
As previously reported (Garza-Brenner et al., 2017), the G allele had the highest frequency in the rs43696138 marker, and the A allele had the highest frequency in the rs109576799 marker in the three populations evaluated. However, the association results obtained in the present study contrast with those previously obtained in Charolais cows (Garza-Brenner et al., 2017), where the two SNPs (rs109576799 and rs43696138) were associated with temperament. Sample size could have been an important factor contributing to this result. It has been proposed that when a marker association is discovered in the Gene-SNP/trait, different factors could affect it, such as the genetic background of the animals studied, in this case Bos taurus vs. Bos indicus, comparing Charolais and Angus with the Brangus breed; selection pressure, as each producer in Mexico selects the phenotypic characteristics that they consider appropriated for the breeding and production system; different frequencies in the founder animals; or random genetic drift (Machado et al., 2003).
As described by Garza Brenner et al., 2017, the biological functions of the HTR2A and DRD3 genes have not been previously reported in cattle; however, according to studies in humans, an effect on performance traits is expected (Supek et al., 2014; Banlaki et al., 2015).
Recently, Dos Santos et al. (2017) identified candidate genes for reactivity in Guzerat (Bos indicus) cattle. One of those genes was ZBTB20, a transcription factor with the potential for association with performance traits such as anxiety, and a strong candidate for reactivity in cattle. The authors also found the dopamine receptor 3 (DRD3) gene 70 kb upstream of the ZBTB20 gene. Therefore, it is important to continue studies to establish the role played by the rs109576799 marker evaluated in this work both in the expression of bovine temperament and in parameters of feed efficiency, as well as to study the linkage disequilibrium of this marker with the intronic marker located in the ZBTB20 gene.
In a study by Llonch et al. (2016), the association of temperament and stress response with productivity, feed efficiency, and methane emissions in cattle was evaluated. No significant associations were found between temperament and feed efficiency; however, they observed that a greater response to stress was associated with a reduction in feed intake. Calmer cattle therefore ingest larger amounts of food, which will be reflected in increased growth and fat deposition compared to more temperamental animals. Likewise, in the study performed by Lindholm- Perry et al. (2014), no association was found for the tested markers; however, these authors also analyzed SNPs in BTA6:38-39 Mb, a region they previously associated with average daily gain (ADG) and average daily feed intake (ADFI). Several SNPs in BTA6 were associated with flight speed (P≤0.005). The study of the genetic architecture of complex traits such as temperament and feed efficiency are a major challenge. The use of genomic approaches will allow to elucidate this complexity, identifying gene by gene the ones that have an influence on these traits.
As a final remark, it is important to mention that the number of individuals studied plays an important role in association studies, and having a greater number of individuals of each genotype evaluated allows more accurate and reliable data. Further research on these and related genes could clarify and validate their proposed effect on temperament and feed efficiency.














