Introduction
Cellulose, a major component of plant biomass, is degraded within the rumen by several bacteria (Dias et al., 2017), fungi (Boots et al., 2013) and protozoa (Newbold et al., 2015). This biomass is hydrolyzed to cellobiose and glucose by cellulolytic microorganisms (Guzman et al., 2016); some anaerobic cellulolytic bacterial strains have developed an efficient enzymatic strategy known as cellulosome (Artzi et al., 2016). Anaerobic ruminal mesophilics digest cellulose by adhering to vegetal fiber (Slutzki et al., 2013). Ruminococcus albus and R. flavefaciens use cellulosomes, pili proteins (Rakotoarivonina et al., 2005) and glicocalix containing extracellular polymeric substances (Weimer et al., 2006), and the catalytic part of the cellulosome includes cellulases, xylanases and pectinases that work synergistically to degrade the complex molecules of the cell wall (Artzi et al., 2016). Microorganisms with these characteristics have potential for the industry of cellulosic-biofuel production since the high cost of converting biomass to sugars is the main obstacle for developing this industry (Venkatesh, 2014; Zhivin et al., 2017).
However, relatively few rumen bacteria have been identified as primary degraders of plant fiber (Flint and Bayer, 2008; Flint et al., 2008). Isolation of these microorganisms can provide useful information about diversity of the ruminal habitat, which would allow identification of fibrolytic enzymes with potential use as forage additives.
Fibrobacter and Ruminococcus are the two genera of ruminal bacteria reported with cellulolytic activity (Wilson, 2011), and not all cellulolytic species have been identified so far. However, molecular analyzes are helping to reveal which phylogenetic groups contribute to fiber degradation (Flint et al., 2008). Wang et al. (2011) reported the isolation and characterization of Shigella flexneri, G3 strain in ruminal liquid. This strain efficiently converted sugars from cellulose as carbon source under mesophilic conditions; thus, it is an attractive candidate for obtaining high yields of oligosaccharides from lignocellulosic biomass. Therefore, the objective of this study was to identify and characterize a microorganism isolated from a cow rumen, capable of hydrolyzing cellulose.
Materials and methods
Ethical considerations
The cow used in this study was cared for according to the standards of the Mexican Council on Animal Care (NOM-062-ZOO, 1999).
Isolation of cellulolytic bacteria
This isolation was performed at the Ruminal Microbiology Laboratory, Colegio de Postgraduados, Montecillo, Estado de México. Briefly, 300 mL of fresh ruminal liquid were collected from the middle ventral part of the rumen of a Holstein cow (520 kg body weight and fed on grassland of Lolium perene L.) using a sterile ruminal cannula (Mateo-Sánchez et al., 2002). A sample of filtered ruminal liquid (1 mL) was inoculated in an anaerobic selective media (ASM) (Cobos, 1995).
The culture medium ASM (100 mL) was composed of 47.9 mL distilled water, 30 mL of clarified ruminal fluid [filtrated in a triple gauze and centrifuged at 12,857 x g for 15 min at 4 °C, and sterilized at 121 °C for 15 min at 15 psi in an autoclave (Felisa, FE- 397, Mexico)]. Five milliliters of mineral solution I [6 g K2HPO4 (Sigma, St Louis, MO, USA) in 1,000 mL H2O]. Besides, 5.0 mL of mineral solution II [6 g H2PO4 (Sigma, St Louis, MO, USA), 6 g (NH4).2SO4 (Sigma, St Louis, MO, USA), 12 g NaCl (Sigma, St Louis, MO, USA), 2.45 g MgSO4 (Sigma, St Louis, MO, USA), and 1.6 g CaCl2-H2O (Sigma, St Louis, MO, USA) in 1,000 mL H2O]. Two milliliters of 8% Na2CO3 solution [8 g Na2CO3 (Sigma, St Louis, MO, USA) in 100 mL distilled water] and 2 mL sulfide-cysteine solution [2.5 g L-cysteine (Sigma, St Louis, MO, USA) dissolved in 15 mL NaOH 2N (Meyer, Tlahuac, Mexico City, Mexico). In addition, 2.5 g Na2S-9H2O (Meyer, Tlahuac, Mexico City, Mexico)], 0.2 g tripticase peptone (Biosciences, San Jose, CA, USA), 0.1 g yeast extract (Sigma, St Louis, MO, USA), and 0.1 mL resazurin (Sigma, St Louis, MO, USA). Nine milliliters of sterile ASM were added to sterile tubes (18x150 mm) containing a strip of Whatman paper No. 541 (3x30 mm) (Sigma, St Louis, MO, USA) as a sole carbon source.
After 48 h incubation at 38 ºC, Whatman paper was removed and inoculated in a sterile ASM, and the process was repeated twice following the same incubation conditions. All the procedures were performed under sterile conditions in a biological safety cabinet (Labconco, Purifier Class II model, Kansas City, MO, USA) with 5% CO2. Petri dishes (Thomas Scientific, Swedesboro, NJ, USA) were used to prepare the solid culture media (SCM) with the same components of the ASM; additionally, bacteriological agar (15 g/L) (Sigma, St Louis, MO, USA) and carboxymethylcellulose (0.28 mg/100 mL) (Sigma, St Louis, MO, USA) was added as the sole source of carbon (Cobos, 1995). The SCM were inoculated with the last recovered culture grown in ASM, sealed and incubated at 38ºC for 72 h. Based on visual inspection of colony morphology, such as color, shape and elevation, colonies with similar morphology were selected and transferred to liquid and solid media (eight times).
Inoculation of culture media and examination of samples were carried out in an Anaerobic chamber (Plas Labs, Lansing, MI, USA), and the incubation of culture in an anaerobic jar (2.5 L AnaeroJar, Oxoid, Basingstoke, Hampshire, UK) provided with AnaeroGen (Oxoid, Basingstoke, Hampshire, UK). Gram staining was performed after each procedure, using an optical microscope Axiostar Zeiss (BioMedical Instruments, Zur Schoenen Aussicht, Zoellnitz, Germany). After massive production in liquid media, purity of the culture was verified with samples observed in the optical microscope (Axiostar Zeiss, BioMedical Instruments, Zur Schoenen Aussicht, Zoellnitz, Germany) and Gram stain; subsequently, the pure culture was lyophilized at -50ºC and 0.135 mBar (Labconco Freezone, Kansas, City, MO, USA) and preserved for further use.
Cellulase activity
The lyophilized culture (strain) was activated by adding 0.1 g of the lyophilized in 9.9 mL sterile culture medium (ASM) containing carboximethyl cellulose as the sole carbon source. After 3 h of the culture hydrated, 10-4, 10-5 and 10-6 dilutions were performed and then a cross-stripe seeding was carried out in Petri dishes that contained SCM, using a sterile bacteriological loop and incubating at 38°C for 72 h in an anaerobic environment previously described. After 10 days of growing in solid media (SCM), single colonies were randomly selected and stained with Congo red solution (1 mg/mL) for 15 min. The dye was washed with NaCl 0.1 M solution. Bacteria making haloes around their colonies were isolated as cellulolytic anaerobes.
Biochemical identification
The lyophilized culture was activated as mentioned previously in the liquid media (ASM) containing carboximethyl cellulose as the sole carbon source.
A 1 mL sample of this suspension was grown in ASM media for 48 h at 39ºC, verifying to reach optimal turbidity (1x108 UFC mL) according to the methodology described by Ley de-Coss et al. (2013). After reaching optimal turbidity an API 50CHB/E (BioMérieux, Marcy l’Etoile, France) bulb was resuspended with 1 mL of culture. The carbohydrate fermentation pattern was assessed in an incubation chamber (Riossa EO71, Mexico DF, Mexico), following manufacturer instructions. Test strips were incubated 24 h at 38ºC and results were analyzed using API WEB software (API 50 CHB v.4.0).
Strain identification and nucleotide accession number
Genomic DNA extraction and molecular biology methods were carried out according to Hatfull and Jacobs (2014) protocols. The extracted DNA was used as template for PCR amplification of the 16S r DNA gene, using the primers 8F (5’-AGA GTT TGA TCC TGG CTC AG-3’) and 1492R (5’-GGT TAC CTT GTT ACG ACT T- 3’). PCR was performed according to Valledor et al. (2014). The reaction mixtures (25 µL) contained 5 µL PCR buffer 5x (Promega, Madison, WI, USA), 2.5 mM deoxynucleoside triphosphate (dNTP), 10 pmol/µL forward and reverse primers, 0.035U of Taq DNA Polymerase (GoTaq® DNA, 5U; 0.08 U/µL; Promega, Madison, WI, USA), and 100 ng template DNA. The samples were amplified using a Bio-Rad DNA Engine with the following thermal profile: 95ºC for 2 min, 30 cycles at 95 ºC for 2 min, 57 ºC for 1 min, 72 ºC for 3 min, and finally 75 ºC for 5 min. The PCR-amplified 16S rDNA was purified (Wizard SV gel and PCR Cleand VP-System; Promega, Madison, WI, USA) and its size was verified by low-melting point agarose electrophoresis (EC Maxicell Primo EC 340, Thermo Fisher Scientific, Waltham, MA, USA) using TAE as running buffer. Staining solution SYBR Green (Invitrogen, Carlsbad, CA, USA) was used for UV visualization of DNA, using a KODAK transiluminator (Gel Logic 100 Imaging System, 365 nm; Eastman Kodak Company, Rochester, NY, USA).
Sequencing was performed at Seeds Biotechnology Laboratory, Colegio de Postgraduados, Montecillo, Estado de Mexico. For each reaction, 1.8 µL of Buffer BigDye Terminator v3.1 (Applied Biosystems, Foster City, CA, USA) and 2 µL of each sample were placed in the sequencing plate using a Genetic Analyzer 3130 (Applied Biosystems, Foster City, CA, USA). The BioEdit Sequence Alignment program (v.7.0.9.0) was used to construct the consensus sequences. The nucleotide sequences were compared by BLASTN program (http://www.ncbi.nlm.nih.gov/ BLAST/). Alignment was carried out using ClustalW (MEGA Program, v.6.0). Phylogenetic dendograms were constructed using bootstrap analysis of 5,000 replicates. The 16S rDNA sequence generated from bacterial isolate was deposited into NCBI database under accession number KM094184.
Results
Morphology and cellulolytic activity of the isolated strain
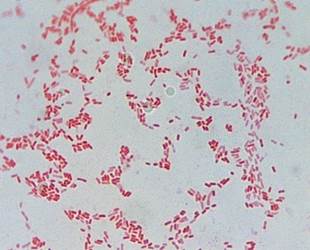
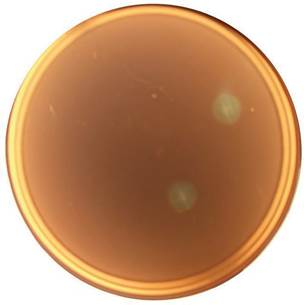
The bacterial strain isolated was a Gram- negative coco bacillus (Figure 1), which forms beige, circular, smooth edges and convex colonies. Bacterial cellulolytic activity based on Congo red staining method was evidenced by the formation of halos (Figure 2) around their colonies due to i β-D- glucanase activity.
Biochemical identification
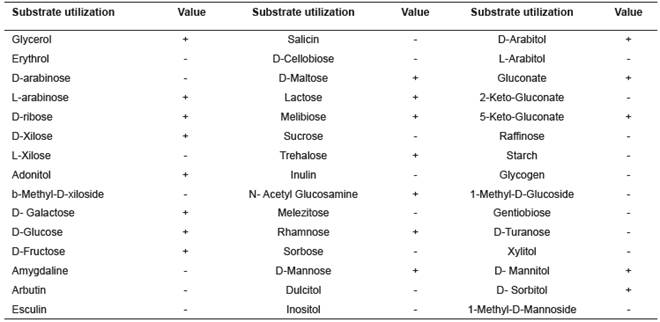
Microorganisms have a fermentation pattern that allow to characterize and identify bacterial genera. The result obtained in our study with the enzimatic gallery API 50 CHB/E was integrated by API WEB Software (Table 1). However, no matching was found in the database.
Phylogeny
The phylogenetic tree based on 16S rDNA sequence is shown in Figure 3. This result allows confirming that the ruminal isolated bacteria belongs to Shigella genus, sharing 98% similarity to other species from this taxon and phylogenetic inference indicates a reliability of 64 with this genus. However, this is an unrecognized species within the genus Shigella, based on the assumption that strains with less than 98% of identity are not related at the bacterial species level.
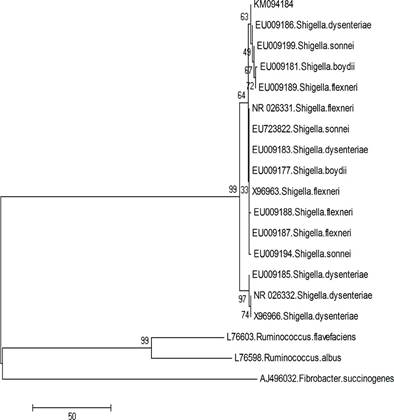
Figure. 3 Phylogenetic tree of the isolated strain and other known Shigella strains based on 16S rDNA gene sequences. The numbers along branches indicate bootstrap values (5,000 replicates). Nucleotide sequence for 16S rRNA of the isolated microorganism is available at NCBI with the access number KM094184.
Discussion
The ruminal cellulolytic bacteria most reported in the literature are Fibrobacter succinogenes, Ruminococcus flavefaciens and Ruminococcus albus (Denman and McSweeney, 2015). There are also reports for Pseudobutyrivibrio, Butyrivibrio, Streptococcus, Enterococcus, Anaerovibrio, Selenomonas, Shaccharofermentans and Actinomyces (Nyonyo et al., 2014). In special cases, like in our study, Shigella genus isolates from the ruminal environment were also described (Akintokun et al., 2014; Baltaci and Adiguzel, 2016; Wang et al., 2011).
Most cellulolytic microorganisms isolated from the rumen are Gram positive (ex. R. flavefaciens and R. albus) (Russell et al., 2009). In contrast, the bacterial strain isolated in this study is a Gram- negative coco bacillus, according with Penatti et al. (2007), of the genus Shigella.
Ruminal microorganisms hydrolyze structural carbohydrates and use monosaccharides and disaccharides for their growth, in addition to other derivatives from fermentation of structural polysaccharides. Shigella sp, isolated from the ruminal ecosystem, releases endoglucanase and xylanase enzymes (Baltaci and Adiguzel, 2016) to metabolize residual cell wall components such as cellulose, cellobiose and xylose (Wang et al., 2011). The presence of halos is an indicator of extracellular production of cellulase (Gupta et al., 2012; Baltaci and Adiguzel, 2016). Endogluconase activity was also observed in bacteria growing in Petri dishes, pressumably because the dye is absorbed by polysacharidic bacterial chains (Ten et al., 2004).
Although API WEB software was not compatible for identifying this microorganism, it evaluated the metabolic activity of the strain, mostly the cellulolytic activity. The ability to ferment glycerol, ribose, D-xilose, sucrose, galactose, glucose, fructose, manose, maltose, mannitol, sorbitol, among others, is in agreement with characteristics reported by Wang et al. (2011) for the genus Shigella.
Few molecular studies have found new bacterial species in the rumen; it is more common to recognize unidentified fungi genera. Many non-identified species are not phenotypically characterized (Tamura et al., 2007). It is also noticeable that many 16S rRNA gene sequences did not show similarity with classified bacteria, which could lead to the development of livestock specific phylotypes.
Molecular identification based on 16S rRNA or 16S rDNA sequencing (Deng et al., 2008; Mosoni et al., 2007) provides valuable information about the presence of new species in different habitats (Ten et al., 2004), particularly in the rumen (Wang et al., 2011). Fonty et al. (2007) reported a microorganism closely related to Shigella boydii (98.2% identitiy, 16S rRNA sequence) in samples isolated from gnotobiotic lambs inoculated with functional ruminal microbiota, and Akintokun et al. (2014) identified species of Shigella genus in ruminal samples from Nigerian breeds of cattle, congruent with our results.
Besides, cellulolytic activity was reported in bacteria of the Shigella genus isolated from the rumen through traditional microbiological processes in culture media selective for cellulolytic ruminal bacteria (Baltaci and Adiguzel, 2016), and pure isolates were identified by phenotypic characterization (colonial and microscopic morphology). This is relevant for further studies about substrate metabolism and the use of genetic techniques that mark a turning point in the analysis of isolated microbial species (Kenters et al., 2011; Creevey et al., 2014).
We conclude that the isolated bacterium is as a member of the genus Shigella, based on the 16S rDNA sequencing, which is in agreement with its ability to hydrolyze structural carbohydrates. Due to the inability of this strain to degrade cellobiose, based on API 50 strips test, additional studies are needed to further characterize the specific mechanisms of cellulose metabolism. The isolated bacterium is an interesting candidate for obtaining oligosaccharides from lignocellulosic biomass.