Introduction
South American rivers have suffered signifi- cant anthropogenic impacts in recent decades. The construction of hydroelectric dams, pollution, and overfishing have strongly affected their natural fish populations (Reynalte-Tataje et al., 2013). To mitigate these impacts, re-stocking programs aimed to release captive-produced progeny can be implemented (Lopera-Barrero et al., 2016). How- ever, these programs may pose genetic risks to wild populations. Mainly, ill-conceived reproduction strategies -such as mating genetically similar individuals- may result in inbreeding (Lopera-Barrero et al., 2014).
Reproductive propagation of captive neotropical fish most often is performed by hormonal induction using two spawning methods: strip or semi-natural. The strip- spawning method consists of hormonal induction, gametes collection by abdominal pressure, incubation, and egg fertilization (Zaniboni-Filho and Nuñer, 2004; Reynalte- Tataje et al., 2013). Semi-natural spawning is characterized by little intervention of the handlers (Lopera-Barrero et al., 2014); after reproductive induction, males and females are placed together in a custom-designed tank with a re-circulation system simulating riverine flow that induces spontaneous spawning, and the eggs are collected by a pipe system that deposits them directly in the incubators (Zaniboni-Filho and Nuñer, 2004; Castro et al., 2019).
Some studies have shown the advantages of semi-natural over strip-spawning on reproductive efficiency and genetic diversity. For example, Zanoni et al. (2016) obtained 87.2 and 8.17% viable eggs for B. orbignyanus in semi-natural and strip- spawning methods, respectively. Zanoni et al. (2019) noted that the percentages of Salminus brasiliensis females who survived after reproduction procedures were 100 and 62.5% for semi-natural and strip-spawning, respectively. Those researchers associated their results with the higher stress suffered by strip- spawned fish. Other studies have suggested that some individuals produce more larvae in a mating than others in some South American species. Doubling male and female numbers in strip-spawning practices could decrease this reproductive dominance (Souza et al., 2018). However, the effect of spawning methods and the number of breeders on the genetic diversity of B. orbignyanus progeny is not completely known.
Therefore, the objective of this study was to evaluate the influence of the reproductive method and number of breeders on the reproductive efficiency and genetic diversity of B. orbignyanus progeny intended for restoration of wild stocks.
Materials and Methods
Ethical considerations
The methodologies used complied with the National Council's Ethical Guidelines for the Control of Animal Experimentation and were approved by the Ethics Committee on the use of animals of the State University of Londrina, Parana, Brazil (CEUA/UEL 17156.2012.50).
Reproductive efficiency
The study was conducted between November and December 2014, the natural reproduction period of this species. A factorial design was used, consisting of two spawning methods (semi-natural or strip-spawning), two different number of breeders (1♀:3♂ and 2♀:6♂) at the same-sex ratio (1♀:3♂), and three replicates. Females (935 ± 0.19 g) and males (915 ± 0.08 g) of B. orbignyanus were selected from the broodstock maintained at AES-Tiete fish farm (Promissao, Sao Paulo, Brazil) based on reproductive traits (abdominal diameter, sperm production and color of the urogenital papilla). The animals were fed a 32% protein commercial extruded diet during the experiment (Guabi nutrition, Orlandia, Sao Paulo, Brazil). Water quality, temperature (26.92 ± 0.94 ºC) and oxygen (5.1 ± 0.12 mg/L) were monitored daily in the breeding tanks.
To induce spawning, females were injected with 5.5 mg kg-1 carp pituitary extract (CPE) in two applications (10% in the first dose, and 90% 12 hours later). Males received 2.5 mg kg-1 in a single dose concurrently with the second dose administered to females (Lopera-Barrero et al., 2014). In strip-spawning, after the second dose of hormonal induction, the breeders were held separately and approximately six hours later caught, their urogenital papilla dried to prevent contamination, and subjected to gentle cephalo-caudal abdominal massage to release oocytes on females and sperm on males. Then, gametes were mixed (50 g of oocytes + 1 mL of semen) in the presence of water (twice the volume of oocytes), and the eggs transferred to incubators (Lopera-Barrero et al., 2014). In semi-natural spawning, after second hormone administration, the breeders were housed in a circular tank with a forced-circulation water system designed to induce semi-natural spawning conditions. Post spawning (about six hours after pituitary injection), the eggs were collected via an outlet pipe at the center of the spawning tank and transferred to incubators (Castro et al., 2019).
Fertilization and hatching rates were calculated in duplicate by counting the number of viable embryos six hours and number of hatched larvae 12 hours after fertilization (n=200) (Godinho, 2007). The percentage of breeder mortality (males and females) at 24 h post-breeding was also recorded.
Genetic diversity
Caudal fin clips (about 1 cm²) of all (n=24) brood fish and 360 (n=90/treatment) 36 h post- hatch larvae were sampled for genetic analysis. Samples were preserved in 70% ethyl alcohol. The DNA of breeders and larvae was extracted using the sodium chloride and alcohol-based protocol described by Lopera-Barrero et al. (2008). After extraction, DNA was quantified in a PICODROP® spectrophotometer (Picodrop Limited, Hinxton, United Kingdom), and standardized to 10 ng/µL final concentration. DNA integrity was assessed in 1% agarose gel, stained with SYBR Safe™ DNA Gel Stain (Invitrogen, Carlsbad, CA, USA).
Eight heterologous microsatellite loci were evaluated: two (BoM5, BoM13) described for Brycon opalinus (Barroso et al., 2003), five (Bh5, Bh6, Bh8, Bh13, Bh16) described for Brycon hilarii (Sanches and Galeti Jr., 2006), and one (Bc48-10) described for Brycon insignis (Matsumoto and Hilsdorf, 2009). Amplifications were performed in a thermocycler Veriti® (Applied Biosystems®) using Platinum Taq DNA Polymerase (Invitrogen, Carlsbad CA, USA) for a 15 μL final reaction volume, with 1X buffer Tris-KCl, 2.0 mM MgCl2, 0.8 μM of each primer (forward and reverse), 0.4 mM of each dNTP, one unit of Platinum Taq DNA Polymerase and 20 ng of DNA (Invitrogen, Carlsbad, CA, USA), according to Castro et al. (2017). The amplified products were subjected to 10% denaturing polyacrylamide gel electrophoresis (0.5X TBE buffer) for seven hours at 180 V. The gel was then stained with silver nitrate according to Bassam et al. (1991), and photographed on a Gel Doc EZ Imager (BioRad, USA). The number and size of alleles were recorded by Kodak EDAS 290-software using a 50 bp DNA ladder (Invitrogen, Carlsbad, CA, USA).
Statistical analysis
The reproductive efficiency assessed by fer- tilization, hatching and brood fish mortality rate was determined by analysis of variance using a randomized 2x2 factorial design. The variables studied were tested for normal distribution and the averages were compared by Tukey test using the ExpDes package (Ferreira et al., 2018) of the R statistical program. The allele frequency (number and size) was calculated for each locus using the FSTAT 2.9.4 software (Goudet, 2005). The presence of null alleles was tested by the Micro-Checker software (Van Oosterhout et al., 2004). Observed and expected heterozygosity, Hardy-Weinberg equilibrium and Shannon in- dex, were calculated for each locus using the GENEPOP 1.2 software (Raymond and Rousset, 1995). Analysis of molecular variance and Genetic differentiation was conducted with the Arlequin 3.0 software (Excoffier et al., 2005).
Results
Reproductive efficiency
The interaction between spawning method and number of breeders was not significant (p>0.05); thus, only main effects are presented and discussed. The best fertilization and hatching rates (p<0.05) were observed for semi-natural (71.12 and 61.23 %) compared to strip-spawned (53.93 and 40.29 %), whereas the number of breeders (1♀:3♂ or 2♀:6♂) did not affect (p>0.05) these parameters within spawning method (Table 1).
Despite the great difference in female mortality rate between semi-natural (33.33%) and strip- spawned (75.00%), this was not statistically relevant (p>0.05). In contrast, male mortality rates were higher (p<0.05) for strip-spawned. Once again, the number of breeders did not affect the male mortality rates (p>0.05) (Table 1).
Genetic diversity
The eight amplified microsatellite loci produced 30 alleles, ranging from three (Bh5 and Bh13) to four alleles per locus (BoM5, BoM13, Bh6, Bh8, Bh16 and Bc48-10), with size between 80 (Bc48-10) and 225 bp (Bh5). Exclusive alleles were not found. The analysis of allele frequencies showed the maintenance and/or increase of genetic diversity between breeders and progenies in all treatment groups, with both sharing more than 80% of the most frequent alleles. Null alleles were found for six loci (BoM5, BoM13, Bh5, Bh6, Bh8 and Bh16), mainly in BoM13, where the 140 bp allele did not appear in the breeders, but was present in all progenies (Table 2).
Higher values for expected heterozygosity (He) compared to observed heterozygosity (Ho) were found in all groups, except for 1♀:3♂ semi- natural breeders. The highest average of He and Ho values was obtained for 1♀:3♂ strip progeny (0.669) and 1♀:3♂ semi-natural breeders (0.594). Deviation from the Hardy-Weinberg equilibrium (p<0.05) was observed only in 1♀:3♂ strip breeders (Table 3). The Shannon index values ranged from 0.8378 for 1♀:3♂ semi-natural breeders to 1.1877 for 1♀:3♂ strip progenies, with the progenies, always having higher values than their parents. Similarly, the inbreeding coefficient (Fis) ranged from -0.027 for 1♀:3♂ semi-natural breeders to 0.269 for 1♀:3♂ semi- natural progenies (Table 3). Molecular variance analysis showed that most of the variation was within rather than between groups, with low genetic differentiation (Fst), 0.004 to 0.046, among groups (Wright, 1978) (Table 3).
Table 1 Fertilization, hatching, and mortality rates of brood B. orbignyanus subjected to two spawning methods with different number of breeders.
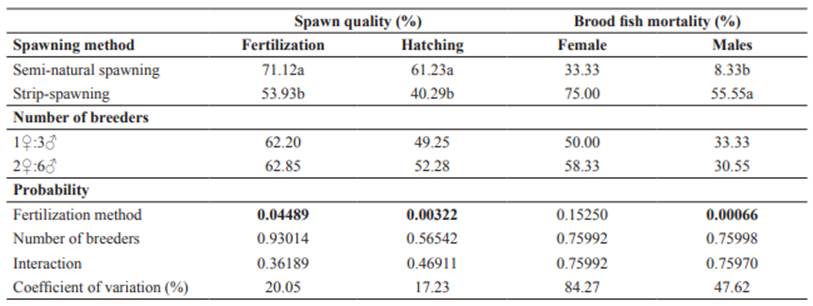
Different letters within columns represent differences (p<0.05) between spawning methods.
Table 2 Number (N), size (S), and allele frequencies of B. orbignyanus subjected to two spawning methods with two different number of breeders.
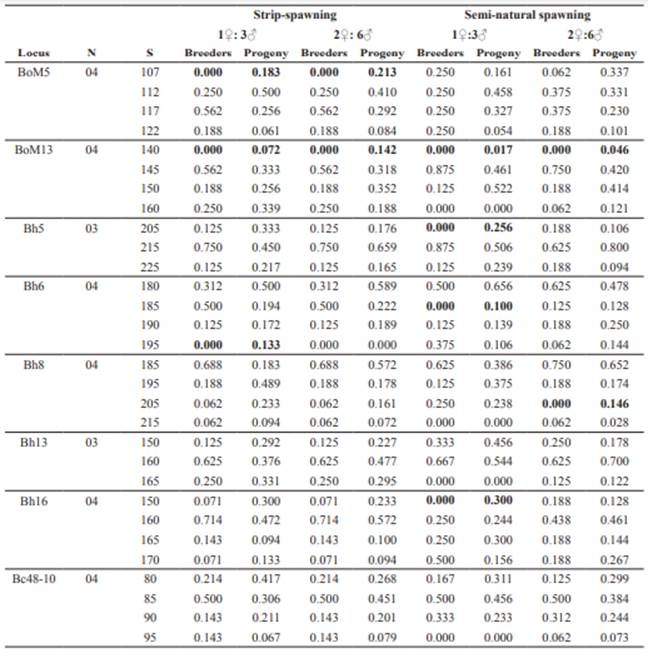
Bold: Null alleles.
Table 3 Observed heterozygosity (Ho), expected heterozygosity (He), Shannon index (H'), inbreeding coefficient (Fis), analysis of molecular variance (AMOVA), and genetic differentiation (Fst) of B. orbignyanus subjected to two spawning methods with a different number of breeders.
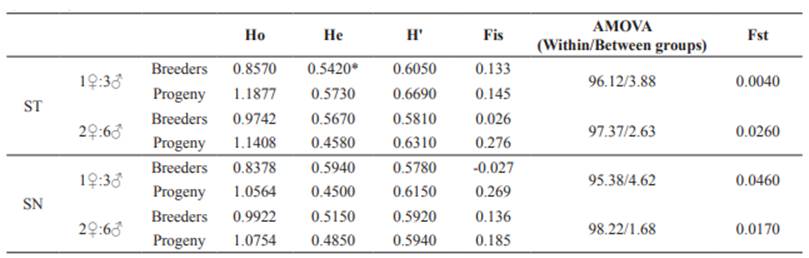
ST: Strip-spawning; SN: Semi-natural spawning. * Significantly different to Hardy-Weinberg equilibrium (p<0.05).
Discussion
Reproductive efficiency
Qualitative spawning parameters obtained during reproductive management of fish can be very variable, mainly due to individual breeding conditions and spawning method. First, the greatest stress suffered by animals subjected to the strip-spawning results in considerable metabolic changes, such as cortisol and glucose levels, as well as hematological parameters, which directly affect gamete quality and breeder mortality (Zanoni et al., 2016). Furthermore, in strip-spawning oocytes can be collected immature or in late to final maturation (Samarin et al., 2015), besides being prone to contamination by feces, blood, or urine (Zaniboni-filho and Nuñer, 2004) that could impair fertilization and hatching rates.
The best fertilization and hatching rates observed for semi-natural spawning in our study is consistent with previous reports. For example, Reynalte-Tataje et al. (2013) working with native Brazilian species, showed that this method had better egg quality than strip-spawned, with 98.7 and 14.7% average fertilization rates, respectively. Zanoni et al. (2019) obtained 91.7% fertilization rate for semi-natural, and only 40.6% for strip- spawning in Salminus brasiliensis. For B. orbignyanus, Zanoni et al. (2016) obtained 87.2 and 8.17% viable eggs in semi-natural and strip-spawning methods, respectively.
The number of breeders did not affect fertilization or hatching rates within spawning method. Improvement was expected, due to the increase in the number of animals may compensate the reduction of the damage caused by the non-participation of animals in the reproductive process, because they are not yet able to reproduce or have poor gamete quality (Castro et al., 2019).
Mortality of breeders during reproductive handling can be detrimental to stock recovery plans and should be avoided, especially when the species is at risk of extinction. Mortality is highly affected by handling stress -especially in sensitive species, such as B. orbignyanus- and injuries caused by stripping -such as loss of scales- which favors fungi and bacteria proliferation leading to animal death (Zanoni et al., 2019; Castro et al., 2019). Sato et al. (1997) attributed 83.3% death of stripped breeders to lesions and hyperemia accompanied with secondary bacterial and fungal infections. We observed high mortality of strip-spawned males. Regarding females, although there was no statistical difference, the strip-spawning method also showed greater mortality; the small number of animals in the 1♀:3♂ groups may have contributed to this lack of significance. Reynalte-Tataje et al. (2013) also found mortality rates similar to those observed in this study, with up to 100% mortality of B. orbignyanus breeders used in strip-spawning. Zanoni et al. (2019) noted that the percentage of Salminus brasiliensis females that survived after reproduction procedures was 100 and 62.5% for semi-natural and strip-spawning, respectively; notably, all the females died 24 h after strip-spawning.
Genetic diversity
The size and number of alleles per locus produced by the eight amplified microsatellite loci were similar to previous studies with the same heterologous markers in B. orbignyanus (Castro et al., 2019). The absence of alleles exclusive to any of the groups suggests there was uniformity of contribution by breeders to progeny across treatments regardless of spawning method (Lopera-Barrero et al., 2014). The presence of null alleles can be related to mutations in the primers' binding region, preventing the annealing during PCR, resulting in unexpected size or no gene product (Chapuis and Estoup, 2007). Often, non- specific microsatellite, such as heterologous, can result in null alleles (Henriques et al., 2017). However, null alleles did not affect heterozygosity in these groups.
The higher values for expected compared to observed heterozygosity in all groups were anticipated considering the dwindling number of the fish in the wild and limited number of breeders available for restocking programs of this fish (Romana-Eguia et al., 2004). In general, the mean values of He and Ho were similar to recent studies with the genus Brycon (Bignardi et al., 2016; Castro et al., 2019).
The deviation from the Hardy-Weinberg equilibrium observed only in the 1♀:3♂ strip- spawned may be common in captive stocks, given the possible genetic drift of these animals, which are isolated and differentiate in these environments (Wahlund effect) or by non- random sampling of animals for reproduction (Romana-Eguia et al., 2004). However, the heterozygous deficit observed indicates a likehood of a decline in genetic diversity in the next generations. Therefore, breeder replacement or acquisition is recommended before this occurs (Souza et al., 2018).
The Shannon index and inbreeding coefficient indicate increase of genetic diversity in the progenies of all treatments. Lopera-Barrero et al. (2010) observed similar results, with increased Shannon index values and expected progeny heterozygosity, that involved individual mating of 1♀:1♂ of B. orbignyanus in semi-natural spawning. On the other hand, analysis of molecular variance and genetic differentiation (Fst) revealed low genetic differentiation between treatments since most of the variation is present within treatments and the Fst had close-to-zero values.
Implications for restocking programs
Genetic monitoring in conservation programs is necessary to optimize reproductive strategies to minimize mortality of broodfish and improve genetic diversity of wild populations. High mortality observed in strip-spawning can reduce future genetic diversity. This is more critical for highly endangered species, which will inevitably force mating between related animals, increasing the inbreeding rate and reducing progeny's survival and adaptability. Genetic analysis, even without differences between treatments -as observed in this study-, are very important for restocking programs since it provides essential information for reproductive management, such as incorporation or replacement of breeders that contribute with adequate genetic variability of progeny. Constant genetic monitoring of the next generations of progeny is strongly recommended. For all these reasons, the results of this study show that semi-natural spawning is the most suitable reproduction method for B. orbignyanus in restocking programs.














