Introduction
A priority for most Latin American countries, including Mexico, is to develop an agricultural sector that is capable of safely and efficiently producing enough food to meet population demands. This objective can be reached through innovation of technologies used to produce bovine embryos, thus allowing to improve the competitiveness of the meat and dairy industry. In vitro embryo production has become an excellent tool to investigate reproduction mechanisms and is used in procedures for gene editing, cloning by somatic cell nuclear transfer (SCNT), and production of embryonic stem cells (Hansen, 2020a). Conventional evaluation of embryos cultured in vitro is based on visual assessment of cell morphology, which may be subjective since it depends on the technician’s experience and judgement. This is reflected in the low pregnancy rates (~50%) recorded for embryos selected under this criterion (Ealy et al., 2019; Hansen, 2020b; Lopes et al., 2020). Additional criteria for embryo quality include: number of cells in the inner cell mass (ICM) and trophectoderm (TE), incidence of apoptosis, eclosion capacity, chromosome abnormalities, and expression of specific genes, among others (Sugimura et al., 2012). Evaluation of these features requires that the embryos are correctly identified, which is challenging to achieve in conventional group (CG) culture since it is conducted with groups of approximately 50 embryos in the same culture dish (Fujita et al., 2006; Salvador et al., 2011). A potential solution to this issue is single-embryo culture; however, several studies (Marianowski et al., 2007; Salvador et al., 2011) have been unsuccessful at single-embryo production because individual embryos do not benefit from the paracrine and autocrine factors secreted by multiple embryos and accumulated in CG culture (Fujita et al., 2006; Gopichandran and Leese, 2006; Paria and Dey, 1990). Therefore, if quality selection is desired, a cell culture system that allows identifying embryos without sacrificing the benefits of CG culture is necessary. A way to achieve this is to sequester single embryos in the same culture medium; and two systems, well-of-the-well (WOW) and polyester mesh (PM), have been used with good results in species such as cattle (Sugimura et al., 2010; Vajta et al., 2000), pigs (Vajta et al., 2008), mice (Komori et al., 2012), and humans (Vajta et al., 2010a; Vajta et al., 2010b). Therefore, the objective of this work was to compare WOW and PM single-embryo culture systems versus CG system as alternatives for generating better- quality bovine embryos.
Materials and Methods
Ethical considerations
All experiments were conducted in accordance with the institutional code for Bioethics Regulation for Animal Welfare of Universidad Autónoma de Chihuahua, México (case number: CFTZyE-Acta-101/2015:ACUERDO 4.2).
In vitro oocyte maturation
Bovine ovaries were collected in a Mexican slaughterhouse (Tipo Inspección Federal -TIF- 366) and transported within 2 hours to the laboratory in sterile sodium chloride 0.15 M (22-25 °C). The cumulus-oocyte complexes (COC) in the ovaries were aspirated from less than 10 mm diameter follicles using a BD 18G x 1
 PrecisionGlide needle (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) with ~50 mmHg pressure of vacuum suction (WOB-L® Dry Vacuum Pumps, Standard-Duty, Welch®, Denver, CO, USA). All chemical compounds used for media culture were from Sigma (St Louis, MO, USA). The chemically defined CDM medium was as follows: 710 mM NaCl, 60 mM KCl, 10 mM KH2PO4, 5 mM Na-citrate, 5 mM NaHCO3, 2 mM CaCl2, 4.9 mM glycine, 1 mM alanyl-glutamine, 20 mM HEPES, 10 mM sodium L-lactate, 0.5 mM Na-pyruvate, 0.5 mM MgSO4, 67 mM non-essential amino acids, and 25 μg/ mL gentamycin. The COC were washed twice in a chemically semi-defined medium for oocyte handling, H-CDM-M [CDM supplemented with 0.5 mM D-fructose, 2.5% fatty acid-free bovine serum albumin (BSA), 22.5 mM NaCl and 20 μg/mL heparin Na salt] and were selected for study if at least three coats of cumulus cells were detected under a Leica MS5 stereomicroscope (Leica Microsystems, Wetzlar, Germany). The COC were cultured in four-well plates (Nunc, Thermo Scientific, Rockford, IL, USA). Briefly, groups of 50 COC per well were maintained in 1 mL of chemically semi-defined medium for in vitro maturation, M-CDM (CDM supplemented with 2 mM D-fructose, 2.77 mM myo-inositol, 0.1 mM taurine, 5% fatty acid free BSA, 15 ng/ mL follicle stimulating hormone (FSH), 1 μg/μL luteinizing hormone (LH), 0.1 μg/μL estradiol- 17β, 50 ng/μL epidermal growth factor (EGF), and 0.1 mM cysteamine) at 38.5 °C with 5% CO2 at 100% humidity for 24 h.
PrecisionGlide needle (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) with ~50 mmHg pressure of vacuum suction (WOB-L® Dry Vacuum Pumps, Standard-Duty, Welch®, Denver, CO, USA). All chemical compounds used for media culture were from Sigma (St Louis, MO, USA). The chemically defined CDM medium was as follows: 710 mM NaCl, 60 mM KCl, 10 mM KH2PO4, 5 mM Na-citrate, 5 mM NaHCO3, 2 mM CaCl2, 4.9 mM glycine, 1 mM alanyl-glutamine, 20 mM HEPES, 10 mM sodium L-lactate, 0.5 mM Na-pyruvate, 0.5 mM MgSO4, 67 mM non-essential amino acids, and 25 μg/ mL gentamycin. The COC were washed twice in a chemically semi-defined medium for oocyte handling, H-CDM-M [CDM supplemented with 0.5 mM D-fructose, 2.5% fatty acid-free bovine serum albumin (BSA), 22.5 mM NaCl and 20 μg/mL heparin Na salt] and were selected for study if at least three coats of cumulus cells were detected under a Leica MS5 stereomicroscope (Leica Microsystems, Wetzlar, Germany). The COC were cultured in four-well plates (Nunc, Thermo Scientific, Rockford, IL, USA). Briefly, groups of 50 COC per well were maintained in 1 mL of chemically semi-defined medium for in vitro maturation, M-CDM (CDM supplemented with 2 mM D-fructose, 2.77 mM myo-inositol, 0.1 mM taurine, 5% fatty acid free BSA, 15 ng/ mL follicle stimulating hormone (FSH), 1 μg/μL luteinizing hormone (LH), 0.1 μg/μL estradiol- 17β, 50 ng/μL epidermal growth factor (EGF), and 0.1 mM cysteamine) at 38.5 °C with 5% CO2 at 100% humidity for 24 h.
In vitro fertilization
After 24 h of COC in vitro maturation, four-well plates (Nunc, Thermo Scientific, Rockford, IL, USA) were prepared, with each well containing 430 μL of chemically defined medium for in vitro fertilization, F-CDM (CDM supplemented with 0.5 mM D-fructose, 2 mM caffeine, 5% BSA and 2 μg/mL heparin and 14 mM NaCl). One mL of purified water was added to the center of each four-well plate. Oocytes at the meiosis II (MII) stage were transferred in groups of 50 to each well. Semen from an Angus bull was used for IVF, as follows: 0.5 mL straws were thawed at 35 °C for 35 s. The semen was centrifuged in a Percoll gradient solution (45 and 90%) at 400 x g for 20 min. The resulting pellet was re-suspended in 4.5 mL F-CDM and centrifuged again (400 x g for 5 min). After the medium was aspirated, the cell concentration was adjusted to 1 × 106 sperm per mL, and 50 μL of this sperm dilution solution was added to each well and the plates were incubated at 38.5 °C in 5% CO2 in humidity-saturated air. Eighteen hours post-IVF, the potential embryos were transferred to 0.5 mL microcentrifuge tubes containing 100 μL of chemically defined medium for handling of early embryos, H-CDM-1 (CDM supplemented with 0.5 mM D-fructose, 2.5% fatty acid free BSA and 22.5 mM NaCl). Then, they were vortexed for 1 min to remove cumulus cells. Then 50 embryos were placed in 500 μL of chemically defined medium for in vitro culture of early embryos, CDM-1 (CDM supplemented with 0.5 mM D-fructose, 2.77 mM myo-inositol, 0.1 mM taurine, 5% BSA, 0.1 mM EDTA and 1 mM NaCl) in each well of a four-well plate. One mL of distilled water was added to the center of the plate. After 60 h at 39 °C in an atmosphere of 5% CO2, 5% O2, and 90% N2 in humidity-saturated air, embryos at the eight-cell stage were incubated and selected in chemically defined medium for handling of late embryos, H-CDM-2 (CDM supplemented with 2 mM fructose, 2.5% BSA, 1.47 mM essential amino acids and 26.5 mM NaCl). Following selection, embryos were transferred to 400 μL of chemically defined medium for in vitro culture of embryos, CDM-2 (CDM supplemented with 2 mM D-fructose, 2.77 mM myo-inositol, 1.47 mM essential amino acids, 5% BSA and 5 mM NaCl) in four-well plates. One mL of purified water was added to the center hole of the plate and it was then incubated with 5% CO2, 5% O2, and 90% N2 in humidity-saturated air at 39 °C for four days. Subsequently, 8-cell embryos were selected and maintained in H-CDM-2 medium and were organized into 20 embryo groups to be cultured in the WOW, PM and CG systems.
Single-embryo culture with shared medium
Two in vitro single-embryo culture systems, PM and WOW, were used with the purpose of generating individual areas to accommodate an embryo with an approximate diameter of 75
 m, and with a 165-
m, and with a 165-
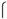 m separation between them, as reported by Gopichandran and Leese (2006).
m separation between them, as reported by Gopichandran and Leese (2006).
The PM system was implemented as reported by Somfai et al. (2010), with some modifications. A polyester mesh (07-300/36, Sefar, Heiden, Switzerland), whose interwoven threads generate areas with 136
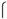 m diameter, each space separated by 200
m diameter, each space separated by 200
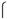 m, were cut into 1.5 mm × 2 mm pieces. These pieces of mesh were placed at the bottom of each well of the four-well dish, and each embryo was placed in the space created by the interwoven threads (Figure 1A).
m, were cut into 1.5 mm × 2 mm pieces. These pieces of mesh were placed at the bottom of each well of the four-well dish, and each embryo was placed in the space created by the interwoven threads (Figure 1A).
The WOW experiments were conducted as reported by Vajta et al. (2008) with some modifications, as follows: in four-well plates containing 400 μl of CDM-2 medium, micro-wells were created at the bottom of the well by applying mechanical pressure using an aggregation needle (DN- 09/B, BLS® Biological Laboratory Equipment Maintenance and Service Ltd., Budapest, Hungary). The resulting micro- wells were 220
 m in diameter, each separated by a distance of 248
m in diameter, each separated by a distance of 248
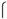 m. Twenty micro-wells were formed in each well of the four-well plate, guided by a template placed on the outside of each well. This template defines an array represented by the letters a, b, c, d and e at the top, and the numbers 1, 2, 3 and 4 on the left side (Figure 1B).
m. Twenty micro-wells were formed in each well of the four-well plate, guided by a template placed on the outside of each well. This template defines an array represented by the letters a, b, c, d and e at the top, and the numbers 1, 2, 3 and 4 on the left side (Figure 1B).
Differential staining: trophectoderm (TE) and inner cell mass (ICM)
Blastocysts were washed three times in 0.1% polyvinylpyrrolidone in phosphate- buffered saline (PBS-PVP) and permeabilized by incubation for 20 s in 0.2% Triton X-100 in PBS-PVP. The blastocysts were then washed three times in PBS-PVP, and transferred to PBS- PVP containing 100 μg/ml propidium iodide to stain the TE cells, incubating them in complete darkness at 37 °C in a humid environment for 5 min. The blastocysts were then washed three times in PBS-PVP. To fix the blastocysts and stain the ICM cells, the embryos were incubated for 30 min in 4% paraformaldehyde (PFA) solution containing 10 μg/ml Hoechst 33258, and finally were washed three times in PBS-PVP.
TUNEL procedure for cell apoptosis detection
Embryos were washed three times in PBS-PVP, then transferred to a 96-well plate containing 100 μl PBS-PVP and 100 μl 4% PFA, and incubated for 60 min at 15-25 °C. Embryos were then washed in another well with 200 μl PBS-PVP. They were then permeabilized by incubation for 5 min in ice (2-8 °C) in a freshly prepared 0.1% Triton X-100 solution in 0.1% sodium citrate, and then washed three times in PBS-PVP. For TUNEL staining of the blastocysts, an In Situ Cell Death Detection Kit Fluorescein (Roche Diagnostics, Indianapolis, IN, USA) was used following the manufacturer’s instructions.
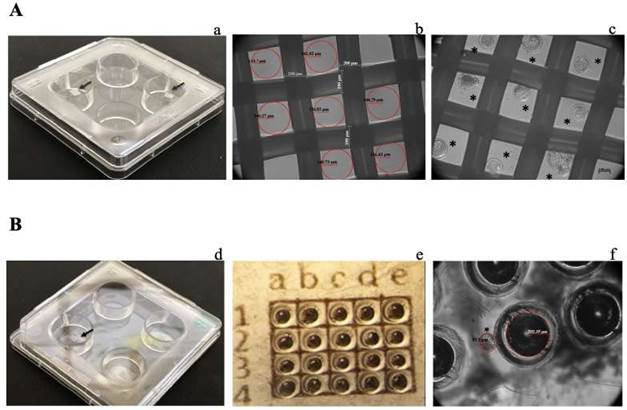
Figure 1 Prototypes of single-embryo cell culture. A. Polyester mesh (PM): a. 4-well dish, the black arrows indicate the micro- wells inside the well; b. View of PM; c. Blastocysts cultured individually in PM. B. Well-of-the-well (WOW): d. 4-well dish, the black arrow indicates the micro-wells inside the well; e. Micro-well template; f. View of micro-wells. In b and f, the red circles indicate the micro-well area. In c and f, * indicates a blastocyst. The images in b, c, e and f were taken with an Axiovert CFL40 microscope (Carl Zeiss, Oberkochen, Germany) using 10× phase contrast.
Permeabilized blastocysts were incubated in the dark in TUNEL reaction solution (1:10 dilution of terminal deoxynucleotidyl transferase enzyme in labeling solution with deoxynucleotidyl triphosphates, dNTPs) for 60 min at 37 ºC. For each TUNEL procedure, positive control embryos were treated with DNase I (3 U/mL) for 10 min at 25 ºC, then subjected to the TUNEL reaction; for the negative control, the embryos were incubated in the absence of the terminal deoxynucleotidyl transferase enzyme.
Blastocyst mounting and microscopy
Blastocysts were placed on a slide in a 10-
 L drop of glycerol and the labeling was observed using an Axio Imager M2 fluorescence microscope (Carl Zeiss, Inc. Göttingen, Germany) at 565 nm for apoptosis, 455 nm for nuclear staining of ICM (Hoechst 33258), and 617 nm for nuclear staining of TE (propidium iodide). Images were acquired using AxioVision Software and AxioCam MRm digital Camera (Carl Zeiss, Inc. Göttingen, Germany).
L drop of glycerol and the labeling was observed using an Axio Imager M2 fluorescence microscope (Carl Zeiss, Inc. Göttingen, Germany) at 565 nm for apoptosis, 455 nm for nuclear staining of ICM (Hoechst 33258), and 617 nm for nuclear staining of TE (propidium iodide). Images were acquired using AxioVision Software and AxioCam MRm digital Camera (Carl Zeiss, Inc. Göttingen, Germany).
Gene expression analysis via qPCR assay
Total RNA of 25 embryos was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, and stored at -80 °C until used for cDNA synthesis. For all samples, the RNA concentration was determined by measuring absorbance at 260 nm using a NanoDrop spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Purity of nucleic acid was determined by calculating the ratio of absorbance between 260 and 280 nm. The cDNA synthesis was performed using a High-Capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions using a mixture of 2 µg of RNA. The reactions were placed in a thermocycler (Corbett Research, San Francisco, CA, USA) under the following program: 60 min at 37 °C, 5 min at 95 °C to deactivate the enzyme, and kept at 4 °C until use. The concentration of obtained cDNA was determined by measurement of absorbance at 260 nm using a NanoDrop spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific, Inc., Wilmington, DE, USA), and the purity of the nucleic acid was determined by calculating the ratio of absorbance between 260 and 280 nm. The cDNA was stored at -20°C until further use. For the analysis of gene expression, all reactions were performed using Real Time StepOne equipment (Applied Biosystems, Carslsbad, CA, USA). Amplification reagents for specific genes were obtained by using the TaqMan Universal Master Mix II and TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA) for POUF5F1 (Bt03223846_g1), ATP5B (Bt03216727_ m1), ID2 (Bt03220879_m1), MATER (Bt03218033_m1), GNAS (Bt03251812_g1), TJP3 (Bt03237880_m1), TP53 (Bt03223222_m19) and CLDN4 (Bt04318530_s1), including the FAM reporter for the genes quantified. The reactions were performed according to the manufacturer’s protocol; each reaction contained 50 ng cDNA, and the quantification of the expression of these genes was normalized to the endogenous GADPH gene (Bt3210913_g1); the results show relative abundance, calculated according to the 2-ΔCt method, where Ct was generated by subtracting the reference gene Ct value from that of the target gene (Livak and Schmittgen, 2001).
Statistical analysis
All statistical analyses were performed using XLSTAT Software (Addinsoft, New York, NY, USA). The data for degenerated and arrested embryos, morulae, blastocysts, TE and ICM are expressed as percentage values. The gene expression data are given as 2-ΔCt relative abundance. The normality of the data was confirmed with the Jarque-Bera test, and non- normal data were transformed with BOX-COX. Normally distributed data were subjected to one-way ANOVA and means comparison was obtained by Tukey’s test. The level of statistical significance was set at p<0.05. All values are presented as means with their corresponding standard error.
To analyze the relationship between culture system and number of cells, type of embryo, and gene expression results, the Spearman correlation was applied, followed by a Principal Component Analysis (PCA), where the principal components (PC) were rotated with the Equamax method and the results shown in a biplot graphic.
Results
Comparison between culture systems
Figure 2 compares the WOW and PM single- embryo culture systems against the CG culture in terms of embryo development. The percentage of blastocysts was similar in all systems (41 ± 4.35% in WOW, 35 ± 3.65% in PM and 36 ± 6.0% in CG; p > 0.05). The same was observed for the embryos that reached the morula stage (29 ± 5.76% in WOW, 26 ± 5.5% in PM and 27 ± 6.7% in CG; p>0.05). The percentage of arrested embryos was similar in all systems (24 ± 6.2% in PM, 20 ± 4.11% in WOW and 20 ± 4.13% in CG; p>0.05). The production of degenerated embryos did show a significant difference; the WOW system produced fewer degenerated embryos compared to the other two methods (11 ± 0.54% versus 15 ± 1.8% in CG and 17 ± 2.7% in PM; p<0.05).
Effect of culture system on blastocyst cell numbers
To examine the effect of culture method on cellular composition of the embryo, we counted the number of cells in the ICM and TE, and the number of apoptotic cells (Table 1). Embryos cultured in all systems had similar number of total cells (81.3 ± 4.0 in PM, 74.4 ± 4.5 in WOW, 68.8 ± 3.6 in CG, p>0.05). Embryos in the three culture systems had similar percentage of ICM (41.4 ± 1.7 in GC, 36.3 ± 1.3 in PM and 38.8 ± 1.8 in WOW, p>0.05). Embryos in the PM system had a significantly higher proportion of TE cells (63.7 ± 3.1%) compared to those in the CG system (58.6 ± 2.6%, p<0.05), but not compared to the WOW system (61 ± 3.2%, p>0.05). The ICM/TE ratio was very close in all systems (0.7 ± 0.06 in GC and WOW, and 0.6 ± 0.03 in PM).
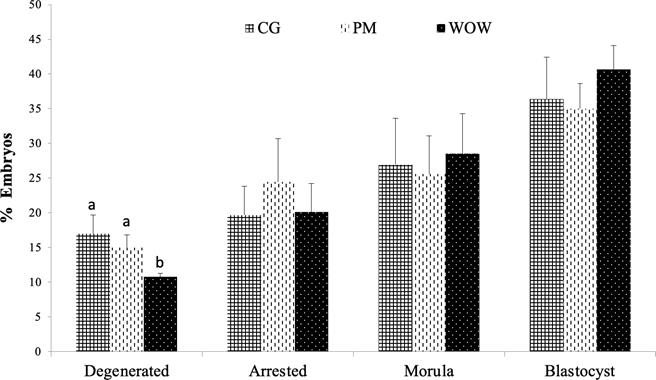
Figure 2 Comparison between the type of embryo developed as of day 7 post-IVF in each culture type: conventional culture in groups (CG), polyester mesh (PM), and well-of-the-well (WOW). Data are shown as means standard error of the percentage of analyzed embryos. In total, 345 embryos distributed in six independent repetitions were evaluated for the three culture types (CG = 117, PM = 115 and WOW = 113).
Regarding the formation of apoptotic cells, embryos in the three culture systems presented very similar numbers, with approximately 2 apoptotic cells for each blastocyst evaluated.
Effect of culture system on relative expression of embryonic genes
The relative mRNA expression levels of embryonic genes POUF5F1, GNAS and TP53 did not show significant differences between WOW, PM and CG culture systems. However, expression of ATP5B gene was higher (p<0.05) in WOW embryos than in PM embryos. On the other hand, expression of TPJ3 gene was higher in PM system compared to CG and WOW systems (p<0.05). Likewise, gene expression of ID2 and CLDN4 was higher in WOW culture than in PM and CG systems (p<0.05). Regarding the expression of maternally expressed MATER gene (a negative control), it was negative in all three culture systems (Figure 3).
Correlation between cell culture system, type of embryo developed, apoptosis, and embryonic gene expression
Principal component (PC) analysis was used to investigate relationships between cell culture systems and the various embryo parameters studied, and two PCs were obtained: PC1 absorbed 53.16% of the variables, while PC2 absorbed 46.84%; together they absorbed 100% of the variables.
Table 1 Effect of the culture system on the number of cells in blastocysts.
| Number of cells per blastocyst | ||||||
|---|---|---|---|---|---|---|
| Culture system | Blastocysts tested | N* | ICM (%) | TE (%) | ICM/TE | Apoptotic (%) |
| 68.8 ± 3.6 | 28.5 (41.4 ± 1.7) | 40.4 (58.6 ± 2.6)a | 0.7 ± 0.06 | 2.2 (3.2 ± 0.6) | ||
| PM | 23 | 81.3 ± 4.0 | 29.5 (36.3 ± 1.3) | 51.7 (63.7 ± 3.1)b | 0.6 ± 0.03 | 2.0 (2.5 ± 0.44) |
| WOW | 26 | 74.4 ± 4.5 | 28.8 (38.8 ±1.8) | 45.6 (61 ± 3.2)ab | 0.7 ± 0.04 | 2.2 (3 ± 0.46) |
The data represents means ± standard error. *From 5 independent repetitions (each with 4 to 6 blastocysts).
a, bValues with different superscripts letters within the TE column differ significantly (p<0.05). ICM: internal cell mass. TE: trophectoderm.
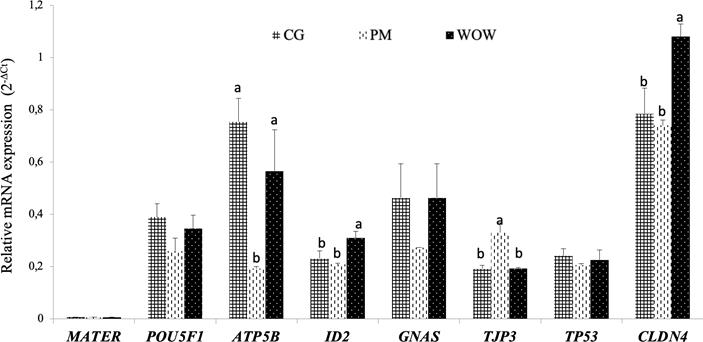
Figure 3 Effect of culture system on relative expression of MATER, POU5F1, APM5B, ID2, GNAS, TJP3 and CLDN4 genes. Data are presented as means ± standard error of the 2-ΔCt value of three independent experiments (each with 25 embryos). For each gene, bars with different letters differ significantly (p<0.05).
This means that enough information was generated to interpret the most important aspects of the data set. The CG system was projected near degenerated embryos and expression of MATER gene; the PM system was projected near arrested embryos, higher values of ICM and TE, and expression of TJP3 gene; and the WOW system was projected toward blastocysts and morulae, as well as toward the expression of CLDN4, ID2 and GNAS genes (Figure 4).
Discussion
The most efficient way for in vitro embryo generation is by group culture (CG), but this approach makes it difficult to label or identify individual embryos, and a sudden movement of the culture dish can cause an embryo to be displaced from its original location. Here, we compared the alternative WOW and PM single- embryo culture protocols reported by Vajta et al. (2008) and Somfai et al. (2010), respectively, where embryos are maintained in defined areas but within the same cell culture medium, to the CG culture system for development of bovine embryos.
Initially, we found that all systems developed similar blastocyst yields. This is interesting because either WOW or PM single-embryo cultures have the ability to generate the same yields that conventional group culture. The WOW system allows each embryo to develop and maintain its own microenvironment, but still sharing autocrine and/or paracrine factors secreted by neighboring embryos (Dai et al., 2012). In addition to this, the partial opening to each micro-well allows for the provision of nutrients and dilution of toxic factors such as ammonia and free radicals (Vajta et al., 2008). The PM system allows embryos to stay in the same microenvironment, and the opening of polyester mesh sections allow the free pass of autocrine and/or paracrine factors (Somfai et al., 2010).
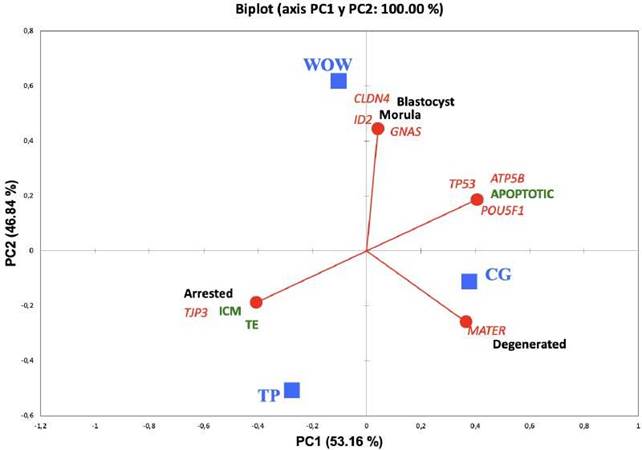
Figure 4 Relationships between culture system and class of embryos developed, type of blastocyst cells, and genes expressed 7 days post-IVF. Blue squares represent three different culture systems (CG, PM and WOW) and circles are the variables: black text designates class of embryos (blastocyst, morula, arrested and degenerated); green text designates type of cells (apoptotic, ICM, and TE). Red lines indicate direction of each variable. PC1: principal component 1; PC2: principal component 2.
The percentage of degenerated embryos was lower in WOW compared to PM and CG systems. Although WOW system yielded the highest percentage of blastocysts, this does not mean that those blastocysts had excellent cell quality. Blastocysts comprise two different cell populations, the trophectoderm (TE) and the internal cell mass (ICM). The TE gives rise to placenta and embryonic membrane development, whereas ICM, through differentiation, generates all the tissues comprising the fetus (Fouladi- Nashta et al., 2005). Thus, prior to implantation, a bovine blastocyst must contain 24 to 34 ICM cells, 83 to 91 TE cells and a minimum of 2 apoptotic cells (Fouladi-Nashta et al., 2005; Thouas et al., 2001). The blastocysts obtained in the three evaluated systems exhibited an adequate amount of ICM and apoptotic cells; in regards to TE cells, the blastocysts obtained in PM showed higher number (p>0.05) of these cells compared to CG and WOW systems. A lower number of TE cells during early embryogenesis has been related with detrimental placental development (Maylem et al., 2017; Meo et al., 2007). An increase of TE cells has a positive effect on pregnancy establishment (Lopera-Vasquez et al., 2017). This supposes an advantage of PM over WOW and GC systems.
The type of in vitro culture is the most critical factor in producing bovine embryos (Corcoran et al., 2006), since it has a direct impact on the phenotype developed by an embryo, and is closely related to the successful development of a blastocyst viable for transfer (El-Aziz et al., 2016). A phenotype modification means a shift in the expression of genes and proteins in early embryos leading to changes in fetal growth, physiological and endocrine parameters after born that affect psychosocial and psychomotor development during life (Sunde, 2019). In ruminants, in vitro embryo culture protocols have been related with alterations in phenotype, such as increased birth weight, gestational length, perinatal death, and large offspring syndrome (Sunde, 2019). In cattle, in vitro production of embryos is influenced by breed; Bos indicus generates greater blastocyst yield than Bos taurus cows (GuimarÃes et al., 2020).
Additionaly, Bos indicus embryos are more tolerant to heat shock (41 °C) than Bos taurus embryos (Monteiro et al., 2018; Paula-Lopes et al., 2013). However, in comparison with Bos taurus, Bos indicus embryos have greater amount of reactive oxygen species (ROS), and this is exacerbated after the cryopreservation process, which causes cell damage and affects embryo viability (Lopez-Damian et al., 2020). Cell culture conditions may negatively affect gene expression profiles and imprinting patterns, even when a blastocyst appears to be successfully developing, based on its cleavage rate and morphological appearance (Khosla et al., 2001). The environment generated is key for embryos to regulate the expression of genes that allow phenomena such as first cell divisions, activation of embryonic genome, morula compaction, and blastocyst formation and expansion (Adjaye et al., 2007; Kues et al., 2008). To ascertain which culture system had the highest amount of embryonic gene expression, we quantified the relative mRNA levels of genes POUF5F1, ATP5B, ID2, GNAS, TJP3, TP53 and CLDN4, using the null expression of maternal mRNA MATER as negative control.
The capacity of blastocysts to direct differentiation of two types of cell lineage, ICM and TE, is regulated by a precise molecular control that involves activation of specific genes for each lineage (Guo et al., 2010). Thus, in most mammalian species, such as human, mouse, porcine and bovine, formation and pluripotent maintenance of ICM is correlated with the expression of POUF5F1 transcription factor (Kirchhof et al., 2000; Schiffmacher and Keefer, 2013; Van Eijk et al., 1999); however, in bovine and porcine models, the expression of POUF5F1 is also correlated with TE, but at a lower rate than for ICM (Kirchhof et al., 2000). Therefore, expression of POUF5F1 gene is indispensable for cell differentiation in the blastocyst (Roberts et al., 2004). Another gene related to TE cell phenotype is ID-2, whose expression is required for maintenance and differentiation of these cells (Garcia et al., 2017) and is key for blastocyst implantation (Roberts et al., 2004; Xie et al., 2013). In the present study, we found neither a significant difference nor a statistical trend for POUF5F1 expression across the three culture systems. This implies that embryos obtained from WOW and PM are very similar to those developed in CG system in terms of POUF5F1 expression; therefore, we can be confident that embryos produced by single-embryo systems have proper differentiation of ICM and TE cell lineage. In the case of ID2 gene, the relative expression of mRNA was higher (p<0.05) in WOW than in PM and CG systems; this suggests that blastocysts generated in WOW may have higher implantation capacity. The biggest challenge of in vitro embryo production is low pregnancy rate. The percentage of post- implantation embryo loss is 70-80% (Corcoran et al., 2006; Goovaerts et al., 2010; Scanavez et al., 2013), thus it is possible that WOW could help improve this embryo wastage.
The environment created in in vitro culture systems may alter genomic imprinting patterns, with deleterious consequences for embryo development (Thurston et al., 2008). After fertilization, epigenetic reprogramming generates DNA methylation patterns that are necessary for activation or silencing of specific genes to drive normal embryonic development (Urrego et al., 2014). Specifically, largescale genome demethylation occurs after fertilization, where most methyl groups -except those in the imprinting control regions (ICRs)- are eliminated from the DNA before the morula stage. Subsequently, de novo methylation occurs in the blastocyst, coinciding with ICM and TE differentiation, where ICM is hypermethylated while TE is hypomethylated (Piedrahita, 2011). The GNAS isanimprinting gene locatedinan ICR. It codes for the subunit of guanine nucleotide- binding protein Gs, which stimulates adenylate cyclase activation after hormonal stimulation and also cyclic adenosine monophosphate (cAMP) production for downstream cellular signal transduction pathways (Khatib, 2004; Sikora et al., 2011). In bovines, GNAS relative expression is maintained at a low level from 2-cell until morula stage and then it increases to high expression levels in blastocyst stage (Jiang et al., 2015; Ruddock et al., 2004). In the present study, we found that relative GNAS expression was not significantly different among the three culture systems, although its expression appeared to be consistently lower in PM. This could imply that the PM system creates a more detrimental environment for GNAS expression; if so, this system could have a negative influence on the genomic imprinting process.
A key point for embryonic development is the energy contribution by mitochondria, which, by means of oxidative phosphorylation, generate most of the ATP required by the growing embryo (Roth, 2018). Gene ATP5B codes for a subunit of the F1 component of ATP synthase, comprising complex V of the oxidative phosphorylation machinery in the electron carrier chain in the mitochondrial membrane (Bougarn et al., 2011; Gad et al., 2012). Our results show that the expression of this gene was lower in PM than in WOW and CG systems; this suggests that embryos may be affected in their mitochondrial metabolism in PM, which could lead to diminished energy capacity.
Apoptosis is considered a normal event in early embryogenesis, necessary to eliminate abnormal cells that may compromise embryo viability (Betts and King, 2001). During bovine embryonic development there is an initial wave of apoptosis at the 9- to 16-cell stage, which later decreases in morula stage and resumes in blastocyst stage (Byrne et al., 1999). Damaged embryo DNA can be reflected by increased expression of TP53 gene, whose protein (p53) acts as a transcription factor to activate expression of the apoptotic gene BAX. In turn, BAX protein acts directly on mitochondrial membrane permeability to liberate cytochrome c, which in turn activates caspase 9, triggering apoptosis (Betts and King, 2001). Thus, in order for an embryo to develop normally it must have an adequate amount of p53 (Liang et al., 2008). The results of the present study show that relative expression of TP53 was very similar in the three culture systems. This indicates that embryos grown in both WOW and PM have adequate expression of TP53, similar to that of embryos cultured in the CG system. This matches the number of apoptotic blastomeres observed by TUNEL.
The first epithelial structure that arises during early embryo development is TE, which covers the blastocyst surface, surrounds the ICM, and generates the blastocoel cavity by transporting ions, water and other small molecules across its epithelium (Fleming et al., 2001). The TE allows the passage of these molecules to the interior of the blastocoel through the action of transmembrane carriers such as Na+K+-ATPase, aquaporins and tight junctions (Furuse and Moriwaki, 2009). In this way, TE works as a barrier isolating ICM from the uterine environment (Moriwaki et al., 2007). Tight junctions become visible around the late morula stage, and in the blastocyst stage they circumscribe TE cells as belts to seal the intercellular space of adjacent cells and create permeability barriers that regulate passage of molecules and ions through a paracellular pathway (González-Mariscal et al., 2011; Moriwaki et al., 2007). Tight junctions are comprised of transmembrane proteins, such as ocludin and claudins, that join inside the cell with peripheral membrane proteins TJP1, TJP2, and TJP3, located toward the cytoplasm, which in turn interact with the actin cytoskeleton and also recruit factors involved in signal transduction and regulation of proliferation and differentiation (Moriwaki et al., 2007). These tight junction transmembrane proteins couple in homo- and/or heterotypic interactions with cognate proteins on the surface of adjacent cells to form the paracellular barrier (Kiener et al., 2007). Tight junctions with claudin-4 appear to seal the intercellular space of TE, where they form aqueous and ion-selective aqueous pores, and contribute to fluid accumulation in the blastocoel cavity, which becomes expanded by elevated hydrostatic pressure (Furuse and Moriwaki, 2009; Serafini et al., 2009). Here, we examined the expression of CLDN4 and TJP3 genes, which code for tight junction proteins. Our results show that CLDN4 expression was significantly higher in WOW, whereas TJP3expression was higher in the PM system. This implies that both single-embryo culture systems have the capacity for developing blastocysts with adequate TE conformation. However, since claudin-4 is necessary for blastocyst expansion, it is possible that blastocysts developed in WOW may have higher capacity for adequate TE generation to allow adequate delimitation of the ICM zone.
In conclusion, compared with the standard CG system, both PM and WOW systems are good options for culturing single, identifiable embryos in the bovine model. This can be considered as an opportunity to improve selectivity of in vitro produced embryos.














