INTRODUCTION
It has been estimated that more than 40% of wood production in Colombia emanates from illegal sources (WWF-Colombia, 2013). In a country where 52% of the area is occupied by natural forests and in which illegal logging is one of the principal drivers of deforestation (IDEAM, 2020), extraction and commercialization of wood generates a significant environmental and socioeconomic impact and is an explicit threat to biodiversity through habitat loss. To guarantee legality of forest products supply chain, it is necessary to obtain official permits and authorizations from Regional Environmental Authorities that enforce national standards that governing the use, management, transport and sale of forest products. In addition, Colombia has been developing wood identification tools for commercial timbers (Covima, 2020; Especies Maderables, 2016; López Camacho et al., 2014; WWF-Colombia, 2013) and implementing policies and initiatives (e.g. Operation Artemisa) that would ensure that forests are protected and wood and wood-derived products are legally sourced. Compliance with and enforcement of international and local laws for legal wood products depend, in part, on the rapid and reliable identification of wood to validate claims of legality (Wiedenhoeft et al., 2019). Such technical or forensic expertise in turn hinges on the design, validation and deployment of robust scientific wood forensic pipelines to identify timber and combat fraud throughout the supply chain (Lowe et al., 2016).
In a typical scenario, when environmental authorities inspect wood in trade, they evaluate a shipment based on a reported commercial or common name used in the region. Unlawful operators typically falsify paperwork, claiming that the wood in a shipment is of lower value species when, in reality, the shipment contains higher-value/endangered and (sometimes) superficially similar species. The only way to detect such a fraud is to inspect a shipment and make provisional identification of the wood to determine if it is consistent with information provided on the paperwork. To make the greatest impact in preventing illegal trade, these inspections of wood must take place in uncontrolled environmental conditions (i.e. at the point of harvest, in a lumber mill and at the port of shipment) in a matter of seconds to establish probable cause for seizure, detention and further forensic analyses, or release the consignment into trade as compliant. If possible, specimens from a detained consignment should be subjected to further forensic analysis in a laboratory using genetic/microscopy/spectral techniques to enable a legally valid identification (Dormontt et al., 2015).
In Colombia, as in most of the world, if any field screening of timber is undertaken at all, then it is human-based, with customs, environmental and police authorities requiring significant training and regular practice in the use of traditional wood anatomical identification methods to achieve a useful level of proficiency. Unfortunately, there is a significant dearth of such human expertise and lack of infrastructure in most control and surveillance posts to perform accurate and reliable identifications. Consequently, field screening of timber more often relies on subjective evaluation of few and often poorly defined characteristics of wood, such as color/appearance, grain/texture, luster and odor-with no reference to anatomical features of the wood. The lack of sufficient human expertise compared to the demand for timber screening is a major bottleneck in ensuring legal timber trade and has established the clear need for reliable field-deployable wood identification technologies (Wiedenhoeft et al., 2019).
Computer vision-based wood identification is an attractive technology for the development of quick, reliable and field-deployable tools for field screening of wood (de Andrade et al., 2020; Filho et al., 2014; Hermanson and Wiedenhoeft, 2011; Khalid et al., 2008; Ravindran et al., 2018; Souza et al., 2020). The availability of affordable, open source, field-deployable, field tested and image-based wood identification platforms (Ravindran et al., 2020) with identification accuracies that match or exceed those of other expensive technologies (Ravindran & Wiedenhoeft, 2020) have further established the potential of computer vision-based systems as effective and scalable field screening technologies, especially in human-mediated contexts.
In this project, we trained a macroscopic image-based identification model of fourteen commercially important Colombian timbers for use in conjunction with the XyloTron system (Ravindran et al., 2020). Our data collection and model development was done as part of a World Wildlife Fund (WWF)-funded international partnership to develop new technology, knowledge and methodologies that can support capacity building for timber tracking and timber forensics in the Colombian timber market. The pilot study described here yields valuable insights into how an international cooperation can develop a deployable solution to a real-world problem on a timescale of months rather than years. To the best of our knowledge, this is the first report on using a computer vision/machine learning model for wood identification in Colombia.
MATERIALS AND METHODS
The transverse surface of specimens from 14 genera (84 taxa, 764 specimens) from xylaria at USDA Forest Products Laboratory (MADw and SJRw), Wood Laboratory of the Universidad Distrital Francisco José de Caldas (BOFw), Tervuren wood collection (Tw) and Xiloteca Dr. Calvino Mainieri, Instituto de Pesquisas Tecnológicas do Estado de São Paulo (BCTw) were polished using progressively finer-grit sandpapers (240, 400, 600, 800, 1000 and 1500) using compressed air and adhesive tape to, as much as possible, remove dust from the cell lumina between each grit. Imaging of polished surfaces was done using the XyloTron (Ravindran et al., 2020), a macroscopic imaging and computer vision system was used for the wood identification. Each image (with dimensions 2048 × 2048 pixels) shows 6.35 × 6.35 mm of tissue. Details of the selected taxa and image dataset are provided in table 1 and Appendix 1, respectively. Species level identification for the selected taxa was not required (and not possible), hence the 4108 images from the 84 taxa were categorized into 14 genus-level classes for machine learning purposes. The image dataset was separated into a training and testing set, such that 80% of the specimens in the dataset contributed images to the training data only, while the remaining 20% of specimens contributed images to the testing set only. The mutual exclusivity between the training and testing data at the specimen level (as opposed to just the image level) prevents the neural network from learning features to identify the individual specimens, thereby making accuracy of the trained model more informative of its real-world wood identification performance.
We trained and evaluated a convolutional neural network (CNN) that comprise a ResNet (He et al., 2016) backbone pre-trained on the ImageNet dataset (Russakovsky et al., 2015) with a custom classifier head using transfer learning (Pan & Yang, 2010) to classify images of the transverse surfaces of wood into 14 genus-level classes. Random patches (with dimensions 2048 × 768 pixels) were extracted from the training set images, down-sampled by a factor of 4 and inputted in the network in mini-batches of size 16. Our data augmentation strategy included horizontal and vertical reflections, rotations and cutout (Devries & Taylor, 2017). The training methodology and hyperparameter settings used were similar to those used in prior works (Ravindran et al., 2019; Ravindran et al., 2020; Ravindran & Wiedenhoeft, 2020).
To evaluate the specimen classification accuracy of the trained CNN, we computed the predicted label of a specimen as the majority label from the predictions on its individual images (Ravindran & Wiedenhoeft, 2020). If, for instance, a specimen of Cedrelinga has 13 images in the test data set and 7 of the images are predicted as Cedrelinga and 6 are predicted as Parkia, the label prediction for the specimen will be Cedrelinga. To arrive at an overall accuracy, the number of correctly predicted specimens was divided by the total number of specimens.
RESULTS
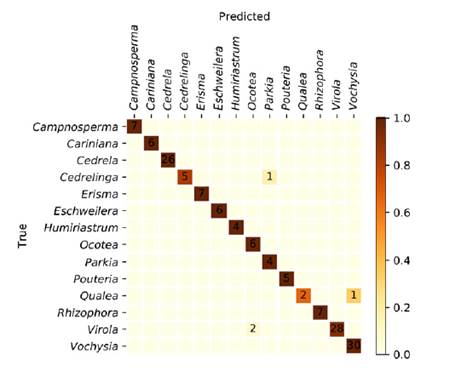
The specimen classification accuracy was 97.3%. The specimen confusion matrix is shown in figure 1, and a total of four specimens were misclassified. The XyloTron, without any optimization for computational performance, requires approximately 2-3 seconds of computational time on a consumer laptop for each image identification, when used in the field.
DISCUSSION
This project developed a highly accurate, machine-learning based wood identification model that can/or should be used to improve timber trade in Colombia. This goal, in the span of less than one year, was established with thanks to a strong international collaboration and partnership. Working together, we were able to build a pilot model in USA, test it in Colombia, improve it with additional data from Colombia’s wood collections and then develop the final model we present here.
The accuracy of the model presented here (figure 1) greatly exceeds the average accuracy and reliability of wood identification experts working in a laboratory setting, as shown in a recent survey conducted in USA (Wiedenhoeft et al., 2019). We are not aware of any country with trained field personnel able to separate these woods with few misclassifications. Only three class-pairs of woods were confused: Cedrelinga-as-Parkia, Qualea-as-Vochysia, and Virola-as-Ocotea.
Cedrelinga and Parkia (figure 2, top) have similar tangential vessel diameter and vessel density and both have vasicentric to lozenge aliform parenchyma. It is interesting to note that a model incorporating UV illumination (Ravindran et al., 2020) would separate this class-pair, since Cedrelinga heartwood is fluorescent and Parkia is not. Qualea and Vochysia (figure 2, middle) have macroscopically similar ray widths and abundance, as well as lozenge aliform to short confluent parenchyma and indeed even in the laboratory. Moreover, using full light microscopic identification, it can be challenging to separate these genera (Quirk, 1980). Virola and Ocotea (Figure 2, bottom) are both somewhat macroscopically similar woods, without any evident characters from the transverse surface to separate the two. If tangential surfaces were added to XyloTron models, the separation of this class-pair would presumably become certain, since Virola has tanniferous tubes in the rays that appear as dark dots on the tangential surface, whereas Ocotea lacks them.

Note that, for each class-pair, the wood anatomical patterns seen with the XyloTron are similar
Figure 2. XyloTron images of the pair (top, middle and bottom) of true class (left) and predicted class (right) for each of the three class-pair misidentifications in the model.
To make better use of the advances from this project, the next logical step would be to conduct a robust field test of the model in Colombia-at markets, lumber yards and at other points of trade control. This would necessitate validation of the XyloTron’s field results by expert human evaluators, ideally by forensic wood identification in the laboratory. It would also be a powerful capacity building exercise for several Colombian colleagues to make a research visit to the Center for Wood Anatomy Research (CWAR) in order to engage in a robust program of advanced training in wood anatomy and machine learning, which are the two sets of expertise needed to adopt and adapt the XyloTron for the Colombian context. With field performance data and increased in-country knowledge of the design, function and machine learning model used by the XyloTron, we could determine the most efficient way to use resources to expand the breadth of Colombian taxa identifiable by the XyloTron.
CONCLUSIONS
With the right partnership, it is possible to develop, implement and complete a machine vision wood identification project with the potential to strengthen control mechanisms for trade and commercialization of wood in the country of origin and within a timescale of months rather than years. This is the first report of such an efficient program of work with a highly accurate machine vision wood identification model as a result. Our identification model had in silico accuracy greater than 97%, an accuracy far higher than any, but by the best forensic experts in a laboratory setting.
Field testing of the model and verification of the model’s results by laboratory testing is a critical next step, ideally followed by a research sabbatical in CWAR to expand knowledge on XyloTron operation and development. With this enhanced capacity, it will be possible to establish best practices for how best to expand the breadth of Colombian woods identifiable by the XyloTron.