Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Colombiana de Química
Print version ISSN 0120-2804
Rev.Colomb.Quim. vol.41 no.3 Bogotá Sept./Dec. 2012
Generation of pharmacophores and classification of drugs using protein-ligand complexes
Generación de farmacóforos y clasificación de drogas utilizando complejos proteína-ligando
Geração de farmacóforos e classificação de fármacos usando-se complexo proteína-ligando
Eliana P. Velásquez1, Neil A. Vásquez2,Pablo A. Gutiérrez1*.
1 Group of microbial biotechnology. School of sciences. Universidad Nacional de Colombia. Medellín, Colombia.
2 Group of animal biotechnology. School of sciences. Universidad Nacional de Colombia. Correspondence concerning this paper should be addressed to Pablo A. Gutiérrez, Medellín, Colombia.paguties@unal.edu.co
Recibido: 14/08/12 - Aceptado: 27/11/12
Abstract
Pharmacophore identification is a very important step in de novo design, lead optimization, chemogenomics, and virtual screening of drugs. Unfortunately, the high cost of comercial software for pharmacophore detection is a common limiting factor for researchers with limited funding. This paper presents a set of freely available perl routines that were designed to help in the process of 3D pharmacophore identification and QSAR studies. These routines also allowed the classification of ligands based on their tridimensional similarity and binding mechanism. The family of phosphodiesterases and their inhibitors were used as test model.
Key words: pharmacophore, inhibitor, protein, enzyme, drug.
Resumen
La identificación de farmacóforos es uno de los pasos más importantes en el diseño de novo, identificación de compuestos líder, quimiogenómica y tamizaje virtual de nuevos medicamentos. Sin embargo, el alto costo de los paquetes comerciales de software para la detección de farmacóforos es un factor limitante para investigadores con recursos limitados. En este artículo se presentan un conjuto de rutinas en Perl que se diseñaron para la identificación de farmacóforos en 3 dimensiones y estudios de QSAR. Estas rutinas también permitieron una clasificación de ligandos basada en su similitud tridimensional y mecanismo de unión. La utilidad de estos programas se probó con los inhibidores de las fosfodiesterasas.
Palabras clave: farmacóforo, inhibidor, proteína, enzima, droga.
Resumo
A identificação de farmacóforos, é um dos passos mais importantes no desenho de novo, identificação de compostos líder, quimiogenômica e triagem virtual de novos medicamentos. No entanto, o alto custo dos pacotes comerciais de software para a detecção de farmacóforos é um fator limitante para os pesquisadores com recursos limitados. Neste artigo apresentamos um conjunto de rotinas em Perl que foram desenhadas para a identificação de farmacóforos em 3 dimensões e estudos de QSAR. Estas rotinas permitiram uma classificação de ligandos baseada na similitude tridimensional e nos mecanismos de união. A utilidade dos programas foi testada com os inibidores da família das fosfodiesterases.
Palavras-chave: farmacóforo, inibidor,proteína, enzima, medicamento.
INTRODUCTION
The pharmacological effect of drugs is generally the result of their interaction with a specific protein target. Compounds with similar activities at the same enzyme or receptor must possess related properties that facilitate their specific binding. A pharmacophore is defined as the 3D arrangement of ligand features responsible for its activity (1, 2). Identification of the pharmacophore is a very important step in de novo design, lead optimization, ADME/TOX studies, chemogenomics, and virtual screening (3-5). The simplest approach for the identification of pharmacophores is based on the alignment of protein-bound ligands and finding their common pharmacophore (1). This method also gives the highest level of resolution as the output consists of a 3D position of an atom associated with its properties (6-8). The performance and applicability of pharmacophore modeling depends on two main factors: the definition and placement of pharmacophoric features and the alignment techniques used to overlay 3D pharmacophore models and small molecules. Identification of the pharmacophore can be a tedious and cumbersome task when many protein-ligand complexes are available.
In this case, it is necessary to superimpose available structures of the protein-ligand complexes. Unfortunately, compounds of different nature can bind to the same protein pocket and, sometimes, the same compound can bind in multiple conformations. For these reasons it is difficult to generalize the chemical features required for binding. Additionaly, the high cost of comercial software for pharmacophore identification can be a limiting factor for researchers with limited funding.
This paper presents a set of freely available Perl routines designed to help in the process of 3D pharmacophore identification and QSAR studies. These routines also allow the classification of ligands based on their tridimensional similarity and binding mechanism. The family of phosphodiesterases (PDE) and their inhibitors was used as test model, which comprises a complex set of ligands that can also bind in different conformations.
MATERIALS AND METHODS
Scripts
All scripts were written in Perl 5.10 (9). Scripts and instructions on how to use them are freely available upon request to the corresponding author.
Data
Comparisons were performed on the following set of phosphodiesterase complexes obtained from the Protein Data Bank.
• PDE3: 1SO2, 1SOJ.
• PDE4: 1PTW, 2QYK, 3I8V, 1RO6, 1RO9, 1ROR, 1TB5, 1XLX, 1XLZ, 1XM4, 1XM6, 1XMU, 1XMY, 1XN0, 1XOS, 1XOT, 1Y2H, 1Y2J, 2QYL, 1MKD, 1OYN, 1Q9M, 1TB7, 1TBB, 1XOM, 1XON, 1XOQ, 1XOR, 1Y2B, 1Y2C, 1Y2D, 1Y2E, 1Y2K, 1ZKN, 2FM0, 2FM5, 2PW3, 2QYN, 3D3P, 3G45, 3G4G, 3G4I, 3G4K, 3G4L, 3G58, 3IAD, 3IAK, 3K4S, 3KKT.
• PDE5: 1UDT, 1UDU, 1UHO, 1T9S, 1TBF, 1XOZ, 1XP0.
• PDE7: 1ZKL.
A list of all ligands and their structures is shown in table 1 and Figure 1.

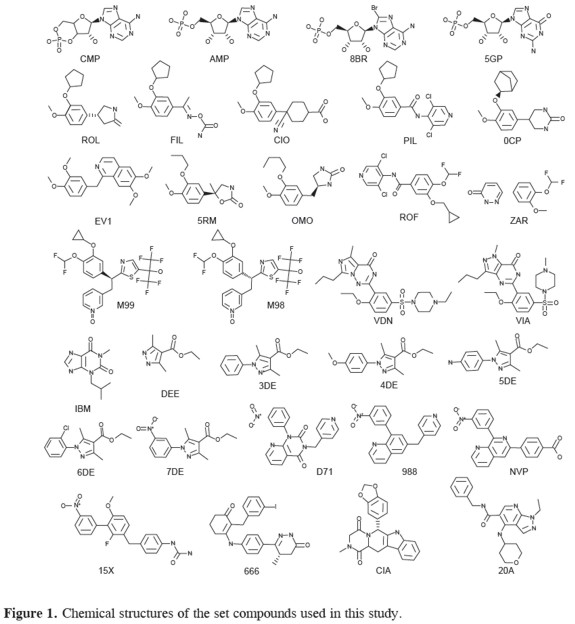
Cluster analysis
Ligands were superimposed with PyMOL (10). Dissimilarity between compounds was measured using the Jaccard distance defined by equation 1.
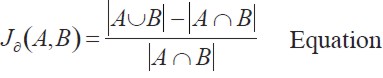 | [1] |
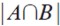 Bcorresponds to the number of atoms closer than 0.7 Åbetween compounds A and B.
Bcorresponds to the number of atoms closer than 0.7 Åbetween compounds A and B. 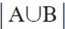 is the total atom count for both compounds. Clustering was done using the Neighbor- Joining method of Nei and Saitou (11) implemented in PHYLIP (12). The tree was drawn using Dendroscope (13).
is the total atom count for both compounds. Clustering was done using the Neighbor- Joining method of Nei and Saitou (11) implemented in PHYLIP (12). The tree was drawn using Dendroscope (13).
Pharmacophore detection and scoring
A total of 27 PDE4D inhibitors were used. The pharmacophore was built by sequentially averaging the position of each pair of atoms closer than a threshold distance of 1.2 Å. The total number of atoms used for each average was written into the temperature factor field of the PDB field. Atom type was assigned to the most frequent atom in the average. Scores were assigned to each inhibitor by adding the temperature field of each matching atom of the pharmacophore.
RESULTS AND DISCUSSION
Phosphodiesterases are a diverse family of enzymes that hydrolyse cyclic nucleotides that play a key role in regulating intracellular levels of the second messengers cAMP and cGMP (14). PDEs are clinical targets for a range of biological disorders, such as congestive heart failure, asthma, chronic obstructive pulmonary disease, depression, retinal degradation, and inflammation (15-17).Currently there is great interest in developing new phosphodiesterase inhibitors with higher selectivity and lower side effects (18). Our set of routines was tested to help understand the binding mechanisms of PDE inhibitors and support the in silico evaluation of potential novel drugs. A flowchart describing the sequential use of these routines is shown in Figure 2.
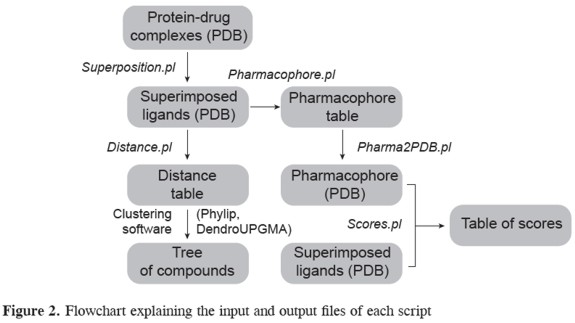
Clustering of compounds The first set of scripts performs the structural alignment of compounds and calculates a distance matrix that can use as input many freely available clustering software such as Phylip or dendroUPGMA (12, 19), (in Figure 3). Scripts were tested using 32 PDE inhibitors from a total of 59 PDB files. However some ligands can bind to the same protein target in two or three alternative conformations (ROL, CIO, ZAR). In total, 66 ligand conformations were analyzed. As a first step, protein complexes must be superimposed. The superposition script, superposition.pl, produces a PyMOL routine that will automatically perform a 3D alignment on the selected structures.
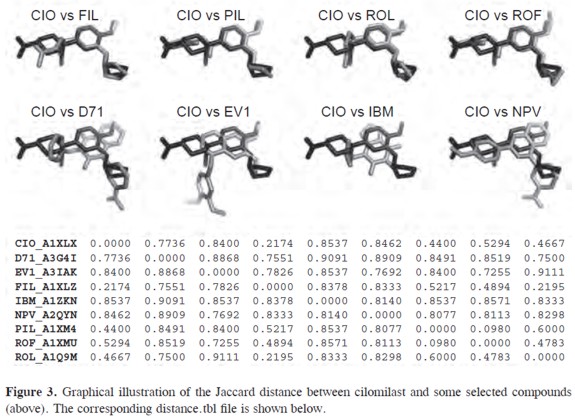
The output will be a set of superimposed ligands in PDB format. All protein information and other non-relevant atoms will be removed at this stage. Output files are named using the compound name, chain id and pdb code from the protein data bank. For example, file ROL_B1OYN_lig.pdb corresponds to the rolipram (ROL) in conformation B as observed in the 1OYN pdb file. Superimposed ligands are used as input for the distance.pl script that will calculate the Jaccard distance for each pair of compounds. A value of zero indicates a complete 3D match correspondence for all atoms present in both ligands. A Jaccard distance of one corresponds to zero 3D matches. A match is defined as a pair of atoms separated by a distances smaller than 0.7 Å. The user can adjust this threshold value. A graphical comparison of Jaccard distance between cilomilast (CIO) and some selected compounds is shown in Figure 3. The output of the distance.pl script will be a square matrix containing the Jaccard distance for each pair of compounds saved as a distance.tbl file (in Figure 3).
Compounds were clustered using the distance.tbl file and the Neighbor-Joining algorithm (11). Nine well-defined sets of compounds, A-H, were obtained (in Figure 4). Cluster A corresponds to rolipram and new generation inhibitors sharing the 4-methoxy-phenyl substructure. Depending on the type of substituents and PDE type, this cluster can be further divided into 5 distinct subsets (A1-A5). It is evident that Rolipram can bind in many different conformations, as reported previously (20). Cluster B group inhibitor NVP bound to diferent phosphodiesterases: PDE4A (NPV_ A2QYN), PDE4B (NPV_A 2QYL) and PDE4D (NPV_A 2QYK). The active sites of PDE4B and PDE4D are mostly comparable. However, PDE4A shows significant displacements of the residues next to the invariant glutamine residue that is critical for substrate and inhibitor binding (21). This difference is clearly seen in tree, were PDEA is an outgroup of compounds PDE4B and PDE4D. Cluster C is comprised by zardaverine and papaverine.
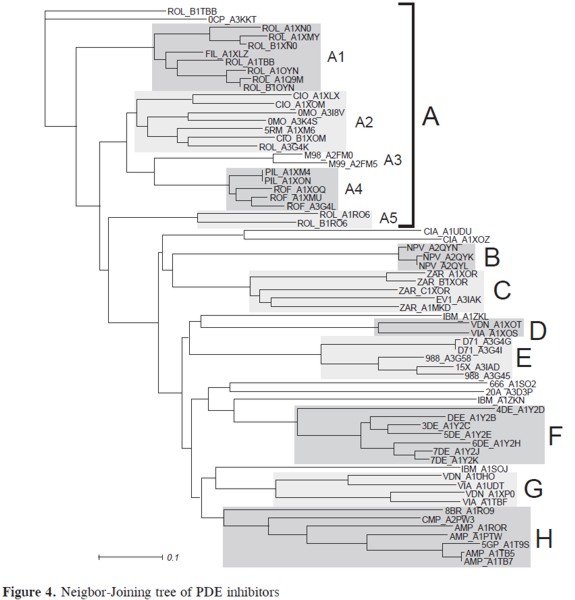
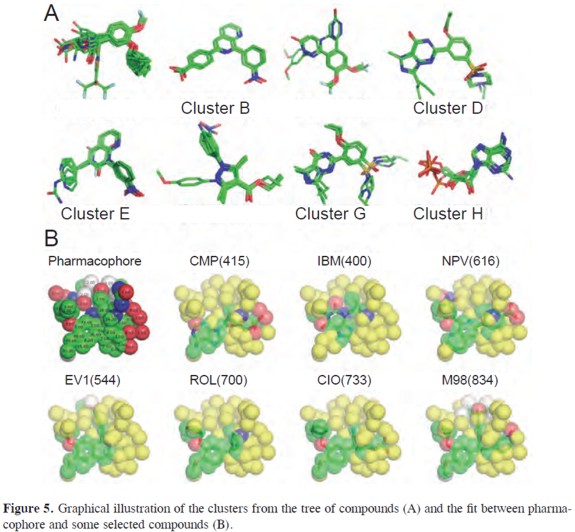
These compounds share the Dialkoxyphenyl group present in rolipram but bind PDEs with lower affinity. These compounds fill only a portion of the active site pocket and lack additional functional groups that can utilize the remaining empty space (22). Clusters D and G group vardenafil (VDN) and sildenafil (VIA). These clusters correspond to complexes with PDE4B and PDE5 respectively. The binding mechanism for these compounds is highly dependent on the type of phosphodiesterase. This is why the same compound belongs to two different clusters. Inhibitors from cluster E include compounds with a 3-nitrophenyl group such as 988, D71 and 15X. In spite of being very similar to NPV, these compounds are bulkier and re- Figure 3. Graphical illustration of the Jaccard distance between cilomilast and some selected compounds (above). The corresponding distance.tbl file is shown below. quire a diferent binding conformation (23). Cluster F comprises compounds 1-7DEE, which have a pyrazole carboxylic ester scaffold with substitutions at three sites (24). Finally, cluster H is composed by nucleotides and analogs. Four compounds did not cluster into any group: tadalafil (CIA), IBMX (IBM), 666 and 20A. An illustration of all clusters is shown in Figure 5.
BUILDING OF PHARMACOPHORES
Three-dimensional (3D) pharmacophore modeling is a technique for describing the interaction of a small molecule ligand with a macromolecular target. These 3D pharmacophores can be used to search for similarities between binding situations or even for similarities between molecules (25). The generation of pharmacophores requires the use of the pharmacophore. pl script that will generate average coordinate for atoms in the vicinity of a threshold value set by the user. In our case a cut-off distance of 1.2 Å gave the most satisfactory results. This file contains an atom count and coordinates for each atom in the pharmacophore. The pharmacophore.tbl file can be converted into a PDB file using the pharma2PDB. pl script. In the resulting PDB file, each atom position is weighted by number of atoms averaged to give the mean their 3D position. This information is recorded under the temperature factor field of the PDB file (9). The atom type field is assigned to the most frequent atom at that position. The resulting pharmacophore for PDE4D inhibitors is presented in Figure 5 together with examples of how this pharmacophore fits some selected compounds. Their pharmacophoric score is shown in parentheses.
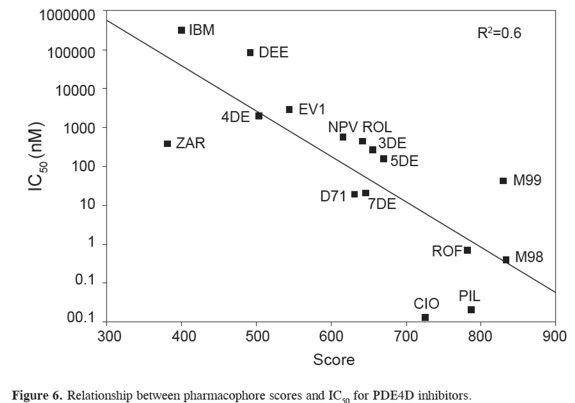
A pharmacophore model has to show predictive power that enables the design of novel chemical structures (26). In order to measure how similar a compound is to the pharmacophore, script score.pl was written which will add the score of each atom of the reference compound. A satisfactory model relating scores with IC50 data has been obtained (R=0.6)(in Figure 6). A closer look at the data shows that zardaverin, piclomilast, and cilomilast have an IC50 at least two order of magnitude lower than the prediction from our scoring scheme. These deviations suggest molecular features that Figure 5. Graphical illustration of the clusters from the tree of compounds (A) and the fit between pharmacophore and some selected compounds (B).greatly improve the binding. Our scorings cheme depends on the total number of atoms averaging to a given position. It is obvious that in some cases strong inhibitors may have some unique substituents that will have low scores in the pharmacophore.The pharmacophore scores can be corrected empirically, by inclusion of new parameters or adjustement of weights, to give a more acurate prediction of inhibition constants. However, these corrections are very case-dependent and correspond to a second phase of QSAR studies, which are beyond the scope of the scripts in this study.
CONCLUSIONS
A set of useful routines for the classification of drugs, 3D pharmacophore identification and preliminary, and QSAR studies has been presented. These programs are freely available and can give an accurate overview of the binding mechanism for large set of compounds. 3D pharmacophore can greatly simplify the molecular features required for inhibitors to interact with a specific protein target. It was also proven that our scoring schemes has predictive power and can be used to help in the de novo design, lead optimization, chemogenomics or virtual screening of novel compounds. The scripts used in this work will be very useful for research groups with limited funding to afford the expensive licenses of commercial software.
ACKNOWLEDGEMENTS
The presentwork was funded by Universidad Nacional de Colombia, Dirección de Investigaciones Medellín (DIME), research grant 90201020.
REREFENCES
1. Podolyan, Y.; Karypis, G. Common pharmacophore identification using frequent clique detection algorithm. J Chem Inf Model. 2009; 49(1): 13-21. [ Links ]
2. Holtje, H. D.; Sippl, W.; Rognan, D.; Folkers, G. Molecular Modeling. 1st ed. Weinheim, Germany: Wiley-VCH; 2008. 310 p. [ Links ]
3. Dror, O.; Schneidman-Duhovny,D.; Inbar, Y.; Nussinov, R.; Wol-fson, H. J. Novel approach for efficient pharmacophore-based virtual screening: method and applications. J Chem Inf Model. 2009; 49(10):2333-43 [ Links ]
4. Tripathi, A.; Surface, J. A.; Kellogg, G. E. Using active site mapping and receptor-based pharmacophore tools: prelude to docking and de novo/fragment-based ligand design. Methods Mol Biol. 2011; 716: 39-54. [ Links ]
5. Klabunde, T. Chemogenomic Approaches to Drug Discovery: Similar Receptors Bind Similar Ligands. Br J Pharmacol 2007; 152: 5-7. [ Links ]
6. Holliday, J.; Willet, P. Using a Genetic Algorithm to Identify Common Structural Features in Sets of Ligands. J Mol Graph Modell 1997; 15: 203-253. [ Links ]
7. Handschuh, S.; Wagener, M.; Gasteiger, J. The Search for the Spatial and Electronic Requirements of a Drug. J Mol Model 2000; 6: 358-378. [ Links ]
8. Finn, P. W.; Kavraki, L. E.; Latombe, J. C.; Motwani, R.; Shelton, C.; Venkatasubramanian, S.; Yao, A. RAPID: Randomized Pharmocophore Indentification for Drug Design. Comp Geom-Theory Appli.1998; 10: 263-272. [ Links ]
9. Tidsell, J. Beginning Perl for Bioinformatics. Sebastopol, USA: O´Reilly; 2001, 368 p. [ Links ] [ Links ]
11. Saitou, N.; Nei, M. The neighborjoining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4(4): 406-25. [ Links ]
12. Felsenstein, J. PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics. 1989. 5: 164-166 [ Links ]
13. Huson, D. H.; Richter, D. C.; Rausch, C.; Dezulian, T.; Franz,M.; Rupp, R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007; 8: 460. [ Links ]
14. Boswell-Smith, V., Spina, D., Page CP. Phosphodiesterase inhibitors. Br J Pharmacol. 2006; 147 Suppl 1: S252-7. [ Links ]
15. Conti, M., Jin, S. L.; Monaco, L.; Repaske, D. R.; Swinnen, J. V. Hormonal regulation of cyclic nucleotide phosphodiesterases. Endocr Rev. 1991; 12(3): 218-34. [ Links ]
16. Teixeira, M. M.; Gristwood, R. W.; Cooper, N.; Hellewell, P. G. Phosphodiesterase (PDE)4 inhibitors: anti-inflammatory drugs of the future? Trends Pharmacol Sci. 1997; 18(5): 164-71. [ Links ]
17. Barnette, M. S.; Underwood, D. C. New phosphodiesterase inhibitors as therapeutics for the treatment of chronic lung disease. Curr Opin Pulm Med. 2000; 6(2): 164-9. [ Links ]
18. Spina, D. The potential of PDE4 inhibitors in asthma or COPD. Curr Opin Investig Drugs. 2000; 1(2): 204-13. [ Links ]
19. Garcia-Vallvé, S.; Palau, J.; Romeu, A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol. 1999; 16(9): 1125-34. [ Links ]
20. Zhao, Y.; Zhang, H. T.; O'Donnell, J. M. Antidepressant-induced increase in high-affinity rolipram binding sites in rat brain: dependence on noradrenergic and serotonergic function. J Pharmacol Exp Ther. 2003; 307(1): 246-53. [ Links ]
21. Wang, H.; Peng, M. S.; Chen, Y.; Geng, J.; Robinson, H.; Houslay, M. D.; Cai, J.; Ke, H. Structures of the four subfamilies of phosphodiesterase- 4 provide insight into the selectivity of their inhibitors. Biochem J. 2007; 408(2): 193-201. [ Links ]
22. Lee, M. E.; Markowitz, J.; Lee, J. O; Lee, H. Crystal structure of phosphodiesterase 4D and inhibitor complex(1). FEBS Lett. 2002; 530(1-3): 53-8. [ Links ]
23. Burgin, A. B.; Magnusson, O. T.; Singh, J.; Witte, P.; Staker, B. L.; Bjornsson, J. M.; Thorsteinsdottir, M.; Hrafnsdottir, S.; Hagen, T.; Kiselyov, A. S.; Stewart, L. J.; Gurney, M. E. Design of phosphodiesterase 4D (PDE4D) allosteric modulators for enhancing cognition with improved safety. Nat Biotechnol. 2010; 28(1): 63-70. [ Links ]
24. Card, G. L.; Blasdel, L.; England, B. P.; Zhang, C.; Suzuki, Y.; Gillette, S.; Fong, D.; Ibrahim, P. N.; Artis, D. R.; Bollag, G.; Milburn, M. V.; Kim, S. H.; Schlessinger, J.; Zhang, K. Y. A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffoldbased drug design. Nat Biotechnol. 2005 Feb; 23(2): 201-7. [ Links ]
25. Schneider, G.; Neidhart, W.; Giller, T.; Schmid, G. "Scaffold- Hopping" by Topological Pharmacophore Search: A Contribution to Virtual Screening. Angew Chem Int Ed Engl. 1999; 38(19): 2894-2896. [ Links ]
26. Dror, O.; Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H. A. A Novel Approach for Efficient Pharmacophore-based Virtual Screening: Method and Applications J Chem Inf Model. 2009; 49(10): 2333-2343. [ Links ]













