Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Acta Agronómica
Print version ISSN 0120-2812
Acta Agron. vol.63 no.2 Palmira Apr./June 2014
https://doi.org/10.15446/acag.v63n2.39879
http://dx.doi.org/10.15446/acag.v63n2.39879
Biotecnología
Segregation analysis of molecular markers in a population derived from Coffea liberica Hiern x C. eugenioides L.
Análisis de segregación de marcadores moleculares en una población derivada de Coffea liberica Hiern x C. eugenioides L.
1Germán Ariel López Gartner*, 2Susan Rutherford McCouch and 3María del Pilar Moncada Botero
1Universidad de Caldas, Calle 65 N° 26-10, Manizales, Caldas, Colombia; 2Susan R. McCouch, Department of Plant Breeding, Emerson Hall, Cornell University, Ithaca, Nueva York 14853, USA; 3Centro Nacional de Investigaciones de Café, Mejoramiento Genético, Cenicafé, Chinchiná, Caldas, Colombia. *Corresponding author: german.lopez@ucaldas.edu.co, Telephone: (57) 68781500 ext. 12189, Fax (57) 68781500 ext. 12194
Rec.: 13.09.2013 Acep.: 27.01.2014
Abstract
Coffee is a very important crop for the world economy. The commercial coffee production is based on two species, Coffea arabica L. (70%) and C. canephora Pierre (30%). C. arabica is the only allotetraploid (2n=4x=44) species, which has its primary center of diversity in the southwestern Ethiopia highlands. C. arabica is the only species cultivated in Colombia and represents a very important economic and social value. However, C. arabica has a very narrow genetic diversity and therefore, the use of diploid species on coffee breeding is very important because they allow to broad its diversity. In this work, the genetic segregation patterns were evaluated on a population of 101 F1 hybrid plants from a cross between the diploid species C. liberica and C. eugenioides using 618 molecular markers, of which 168 SSRs and 2 ESTs exhibited polymorphic patterns that allowed segregation analysis. Approximately 24% of the loci were null alleles, and the segregation distortion reached 23.5% at α < 0.01. A total of four segregant types were observed, from up to seven possibilities. The origins of null alleles, possible causes of segregation distortion, different segregation patterns obtained and recombination frequencies are discussed. The knowledge derived from this study allowed to better understand the genetic segregation behavior of these markers with the aim of developing genetic maps that have important applications for coffee breeding programs as well as other commercial crops.
Key words: Coffee, microsatellites, segregation distortion.
Resumen
El café es un cultivo importante para la economía mundial. La producción comercial de café se basa en dos especies, Coffea arabica L. (70%) y C. canephora Pierre (30%). La primera es la única alotetraploide (2n = 4x = 44) que tiene su centro primario de diversidad en las tierras altas del suroeste de Etiopía; la segunda es la única cultivada en Colombia y representa un importante valor económico y social. Sin embargo, C. arabica tiene una base genética muy estrecha y por ello es importante el uso de especies diploides en el mejoramiento del café para ampliar su diversidad. En este trabajo fueron evaluados los patrones de segregación genética en una población consistente en 101 plantas híbridas F1 de un cruce entre la especie diploide C. liberica y C. eugenioides utilizando 618 marcadores moleculares, de las cuales 168 SSR y 2 EST exhibieron patrones polimórficos que permitieron el análisis de segregación. Aproximadamente 24% de los loci fueron alelos nulos y la distorsión de la segregación alcanzó 23.5% (α < 0.01). Se observaron un total de cuatro tipos segregantes de entre siete posibilidades diferentes. Se discuten los orígenes de los alelos nulos, posibles causas de distorsión de la segregación, diferentes patrones de segregación y frecuencias de recombinación. El conocimiento derivado de este estudio permite entender mejor el comportamiento de la segregación genética de estos marcadores, con el fin de desarrollar mapas genéticos que tienen aplicaciones importantes para los programas de mejoramiento de café y de otras especies comerciales.
Palabras clave: Café, microsatélites, distorsión de segregación.
Introduction
According to the International Coffee Organization (ICO), currently there are some 70 coffee producer countries around the world, of which the exporting members of the ICO are responsible for over 97 percent of world output. In 2010 total coffee sector employment was estimated at about 26 million people in 52 producing countries (ICO, 2014).
Coffee belongs to the genus Coffea, subgenus Coffea (Rubiaceae). The genus includes approximately 80 taxa, most of which are diploids with 2n = 2x = 22. The only tetraploid is C. arabica L., with 2n = 2x = 44 (Bridson and Verdcourt, 1988). The two main cultivated species are C. arabica and C. canephora Pierre. An additional species of coffee that is cultivated less extensively is C. liberica (Van der Vossen, 1985).
For breeding purposes one of the major obstacles is the lack of genetic diversity in the gene pool of C. arabica, making it necessary to use diploid species to introgress genes of agronomic importance (Lahermes et al., 2011). Besides, given the perennial condition of coffee, the implementation of markerassisted selection (MAS), allows the reduction of time required during selection. The use of co-dominant markers, such as SSR (Simple Sequence Repeats), improve the efficiency and accuracy of linkage analysis in diploid species, particularly in those with a high degree of heterozygosity (Jiang and Zeng, 1997) and therefore helps in the implementation of MAS for quantitative traits.
In order to exploit the genome of coffee and bypass the problem of low genetic variability in cultivated species, it is feasible to use its diploid relatives in the construction of a molecular genetic linkage map. Coffe eugenioides is a good candidate to generate a mapping population because of its close evolutionary relationship with the cultivated coffee C. arabica, which is derived from hybridization between C. eugenioides and C. canephora (Lashermes et al., 1999). An additional diploid species of interest is C. liberica, which is known to harbor coffee rust resistance (Prakash et al., 2004) and other traits of economic importance.
However, segregation distortion is a phenomenon of frequent occurrence and wide distribution in plants that has been detected particularly in diverse classes of mapping populations derived from different species (McCouch et al., 1988; Barret et al., 2004). Genetic mapping projects that report segregation distortion propose several different mechanisms to explain this phenomenon. Among the mechanisms that are mentioned with more frequency is the rejection or elimination of male or female gametes during fertilization, the selective fertilization of particular gametic phenotypes and the abortion of zygotes (Xu et al., 1997).
Also it has been suggested that certain loci of the gametophyte can be responsible for a partial or total elimination of gametes carrying certain parental alleles, in which case the distortion of allele segregation is derived from the linkage between the marker alleles and the gametophyte alleles that confer a diminution of the pollination capacity, also known as pollen killer genes (Harushima et al., 1996). This phenomenon has substantial importance for the recovery of specific recombinant genotypes that must be obtained from breeding populations, especially when the populations have been obtained from interspecific crosses, in which the recovery of desirable recombinants is limited due to the nonrandom survival of the descendants (Lashermes et al., 2001).
The main aim of the present study was to analyze the differences in segregation patterns, the genetic distortion of segregant molecular loci in a segregating population derived from the interspecific cross between Coffea liberica x C. eugenioides, as well as to verify the presence of null alleles and the differences in genetic recombination in the two species coffee studied. This segregation analysis was then used to construct a genetic linkage map in the same population (Lopez et al., 2013).
Materials and methods
Plant material
Plant material is maintained in a germplasm bank of the agricultural station of Naranjal at the National Center for Coffee Research (Cenicafé, Chinchiná, Colombia). The segregating population consisted of 101 F1 interspecific hybrid plants developed by cross pollination (CP) between one plant of C. liberica (2n = 2x = 22) as the female parent and one plant of C. eugenioides (2n = 2x = 22) as the male parent. Because these two species are heterozygous, this kind of population is known as a pseudotestcross (Grattapaglia and Sederoff, 1994).
Pollinations were performed by hand, using plastic bags to protect emasculated flowers. Young coffee leaves were packed in wet paper in an isothermic box and brought to the laboratory for immediate DNA extraction. Parental plants were chosen based on gametic compatibility and regular meiotic behavior in interspecific hybrids in order to guarantee a reasonable population size for segregation and linkage analysis. Coffe eugenioides was selected because it was a progenitor species of the cultivated tetraploid C. arabica (Lashermes et al., 1999). Both C. libericai and C. eugenioides share approximately 50% genetic similarity with C. arabica and, between them, share 45% genetic similarity (Moncada and McCouch, 2004). Coffe liberica originated in East Africa while C. eugenioides originated in Central Africa. Both grow mainly in wild habitats, but C. liberica is also commercially cultivated in some countries and used for coffee mixtures. In addition, compared with C. eugenioides, C. liberica has a high content of chlorogenic acids, implicated in the species' organoleptic properties and defense response mechanisms (Ky et al., 1999). Coffe eugenioides has very low caffeine content, 0.2%, when compared with levels in C. liberica that reach 1% (Barre et al., 1997). Additionally, there is evidence that C. liberica has genetic resistance to several biotic and abiotic stresses, including resistance to the coffee Berry borer (CBB) Hypothenemus hampei Ferrari (Romero and Cortina-Guerrero, 2004) and to the coffee leaf rust caused by Hemileia vastatrix Ferrari (1867) (Prakash et al., 2004).
DNA extraction
Genomic DNA extraction for each plant was done starting with 10 g of young apical leaf tissue, according to a protocol of Ky et al. (2000). DNA content and integrity was evaluated by spectrophotometer and by 1% agarose gel electrophoresis, comparing each sample to 2DNA/EcoRI + HindIII standard (Promega). The DNA samples were re-suspended in TE buffer and kept at -20 °C until use.
Molecular markers and polymorphism detection
A total of 618 co-dominant PCR-based molecular markers were evaluated. These included 232 microsatellites (Simple Sequence Repeats or SSRs) developed by Cenicafé from a short insert genomic DNA library of C. arabica 'Caturra' using hybridization-based screening with 26-mer oligonucleotides, (GA)13 and (CA)13, according to the laboratory protocols of McCouch at Cornell University, Ithaca, N.Y. U.S.A. (Unpublished data). Additionally, the rest of the markers were developed for other groups, but intellectual property rights still prevent from publication of the sequences: 315 SSRs by Giorgio Graziosi, University of Trieste, Italy; 20 ESTs (Expressed Sequence Tags) and 51 COS (Conserved Orthologous Sequences) from Solanum lycopersicum (SOL Genomics Network, Cornell University, Ithaca, N.Y. U.S.A.).
The amplification was performed using optimized parameters for each primer pair (Montoya et al., 2006). The amplifications were carried in a total reaction volume of 25 µL. he optimum mix of reagents was 2 µL of DNA (50 ng/µL), 0.75 µL of each primer (diluted to 10 jtmol/L), 2.5 µL of 10x buffer, including 15 mmol of MgCl2, 0.2 mmol/L each of the dNTPs, and 1 U of Taq polymerase. The optimum PCR profile for all markers was an initial denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C for 1 min and extension at 72 °C for 1 min. After 30 cycles, a final incubation at 72 °C for 5 min was allowed. All amplicons were further evaluated on silver-stained 4% polyacrylamide gels.
Data analysis
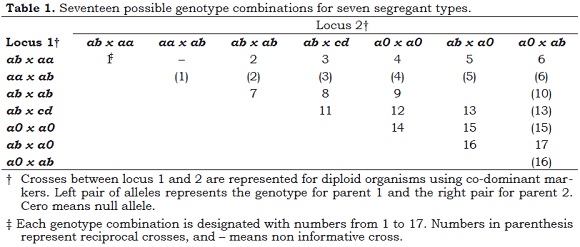
Electrophoreticly visible alleles were designated with lower-case italic letters, while failure to detect alleles after several attempts of PCR was interpreted as evidence of null alleles and designated as 0. For the segregation analysis all possible genotype combinations for codominant markers with mendelian segregation in a diploid organism were considered, as shown in Table 1 (Ritter and Salamini, 1996; Maliepaard et al., 1997).
Segregation analysis was performed after all the individual genotypes were defined for each marker and a database was created. The database was the object of a verification process that included triple gel reading. The segregation ratios of genotypes were tested against expected ratios using the chi-square test. Segregation distortion calculations were estimated by chi-square tests for pairs of linked loci at LOD > 10 taking into the account linkage phase determined by the JoinMap 3.0 software (Van-Ooijen and Voorrips, 2001).
Detection of null alleles
The presence of null alleles was tested in two ways. First, status was based on the phenotype observed among the progeny and their phenotypic and genotypic proportions. Secondly, the presence of null alleles may be due to the lack of correct annealing of the marker primers, even after several attempts at PCR optimization. Nulls were verified nulls by redesigning new primers in such a way as to not overlap with the first set of primers in the original sequences adjacent to each microsatellite locus. This strategy was applied to markers CFGA1254, CFGA205, CFGA55, and CFGA99.
Results and Discussion
A total of 618 molecular markers were evaluated in CP population. It was found that 252 markers were polymorphic; the rest were monomorphic or not scorable for different reasons, including technical difficulties during amplification. From the polymorphic markers, 82 were homozygous in both parents, although they were polymorphic between parents they were not useful for the analysis; the remaining 170 (168 SSRs and 2 ESTs) were used for the segregation analysis.
Segregant genotypic classes.
Configurations found in the segregation analysis of the molecular markers are graphically represented in Figure 1. There are some configurations that do not segregate in the descendants including type vvxvv, monomorphic and monozygotic in both parents; type vvxzz, two alleles but homozygous in both parents and type vvxv0, two alleles but one of them null in the heterozygous parent.
The segregant types considered here are described in the Table 2. Most of the markers analyzed segregate from C. liberica, segregant type 2, followed by those of bi-parental origin, segregant type 4.
Indeed, it is important to note that almost a third of the segregant loci (49 loci or 28.8%) are of the type that provides us with the most genetic information. These types of loci can be used to make genetic inferences for both parents because their segregant types of abxcd or efxeg represent important support for the robustness of the genetic map in these diploid species. In contrast, the segregant type hkxhk had the lowest representation with only 3 loci (1.8%), an observation that is advantageous, considering that this segregant class provides the least amount of genetic information because it implies dominance.
The superior number of segregant markers for C. liberica compared to C. eugenioides could be due to difficulties that arose during the DNA extraction for the latter species, which may have resulted in a decrease in the quality of the template for PCR. The resulting bands associated with the alleles of the male parent were generally less intense, making its visualization difficult and allowing the predominance of informative markers for the female parent.
Segregation distortion
The magnitude of segregation distortion was analyzed for 170 loci with respect to the expected mendelian proportions of segregation following six different statistical criteria; the three most common and practical criteria are discussed here. Using exact significant levels, less markers tend to be distorted at α = 0.01 and α = 0.005; 5 and 8 markers, respectively (Table 3). With the criterion of α < 0.10, segregation distortions were observed for 77 loci, corresponding to 45.3% (Table 3). Segregation distortion at the significant level of α < 0.05 grouped 62 markers or the 36.5% and with α < 0.01 40 markers were grouped, representing 23.5% (Table 3).
No direct relation between segregation distortion and segregant type was observed. Distorted loci were found for all segregant classes in proportion to the number of loci observed for each segregant type. Nevertheless, some tendency towards preferential distortion can be identified for one of the parents in certain regions of a linkage map when it was calculated (López and Moncada, 2005; López et al., 2013). These results support the hypothesis that there are genomic regions that are genetically determined by factors of segregation distortion, and that these control the direction of segregation deviation in favor of a specific progenitor within the linked groups. The evidence suggests that C. eugenioides is the parental species with a greater tendency to present segregation distortion with respect to each segregant type (Table 3, column A). This finding is consistent with other studies that demonstrated that a deviation in segregation arises most commonly by paternal gametophytic selection through selective processes influenced by the gyneceum (Luo et al., 2001).
The distribution patterns as well as the rank of values of the segregation distortion detected here are similar to those obtained in other experiments of segregation analysis and genetic mapping with populations from different species of the Coffea genera. For example, it has been verified in two tetraploids, var. Catimor and var. Mokka, that segregation distortion values ranking from 22% of distorted loci in Catimor to 45% in Mokka are distributed widely in the genomes of these two cultivars (Pearl et al., 2004). In the interspecific map that was constructed in species C. pseudozanguebariae x C. liberica var. Dewevrei, the segregation distortion reached 30% of loci evaluated (Ky et al., 1999). In another population derived from the cross between C. canephora and a double haploid of the same species, proportions of segregation distortion ranking between 13% and 44% (α < 0.05) were found in association with a width distribution of the distorted loci on the genome of this species (Lashermes et al., 2001). In contrast, in a map constructed from a population of double haploids of the diploid coffee C. canephora, a more moderate proportion of segregation distortion was found, varying between 12% for RFLP markers and 20% for RAPD markers, with α < 0.01 for both kinds of markers (Paillard et al., 1996).
Segregation distortion is a common phenomenon and broadly distributed in plants. It has been detected in different mapping population types of different species and by using different types of genetic markers. Several mechanisms have been argued to explain the existence of this phenomenon, including the elimination of some gametes during some stages of the fecundation process, selective fertilization of gametic genotypes and zygotic abortion (Xu et al., 1997). It has also been suggested that some particular loci in the gametophyte could be responsible for the partial or total elimination of male or female gametes carrying incompatibility alleles, in which case, segregation distortions are due to genetic linkage between incompatibility loci and marker loci (Harushima et al., 1996). Segregation distortions should be considered during construction of genetic linkage maps because in some instances it may seriously affect the calculations of the distances among loci.
Null alleles analysis.
Of 170 segregant loci, 41 (24.1%) exhibited the presence of 1 to 3 null alleles per locus that were undetectable using the original PCR primers. Considering each segregant type, of the 41 loci with null alleles, 15 (36.6% from nulls, 53.6% from male parent segregants) exclusively presented the null allele in C. eugenioides, 18 (43.9% from nulls, 20% from the female parent segregants) only in C. liberica and 8 (19.5% from nulls, 4.7% from total segregants) presented the null in both parents. When considering both, the number of null alleles per locus and the segregant type, 22 loci presented 1 null per locus, 6 of them type nnxnp, 4 of type lmxll and 12 of type abxcd or efxeg. Fifteen loci presented 2 nulls per locus, 6 of type nnxnp, 5 of type lmxll and 4 of type abxcd or efxeg. Finally, 4 loci presented 3 nulls per locus, 3 of type nnxnp and 1 of type lmxll. Clearly, proportional to the number of segregant loci for each parent, C. eugenioides exhibits a larger tendency towards the presence of null alleles, an aspect that is perhaps related to the fact that its DNA was recalcitrant to PCR amplification in many cases.
To verify whether, at least in some cases, the null alleles are due to failure by primer sequences to recognize the target DNA during PCR, an additional experiment was conducted in which some primers were redesigned for loci suggesting the existence of null alleles. From the null allele assays for the marker loci CFGA1254, CFGA205, CFGA55 and CFGA99, only the last locus gave some evidence of annealing failure as an explanation for the observation of a null allele (Figure 2, lower right).
Given that the markers used in this study were transferred from the tetraploid C. arabica var. Caturra, this finding suggests that, at least for some marker loci, it is probable that alterations in the sequence of the flanking regions of the microsatellites have occurred during the course of the evolution of this species. Similar observations have been found in other species where detailed sequence analyses demonstrate that base substitutions may explain the presence of null alleles. In rice it has been shown that up to 62% of null alleles are the result of sequence alterations in the flanking regions of the SSRs, caused mainly by point mutations (Chen et al., 2002), a mechanism that can affect important segments of polyploid genomes and the process of differentiation of homologous chromosomes (Eckard, 2001).
Differences in genetic recombination.
Differences between parents with regard to recombination frequencies were studied for 18 map intervals (13.5% from total intervals and 13.5% of genome coverage in cM). These intervals were chosen using markers that provided the most genetic information, corresponding to segregant types abxcd or efxeg, heterozygotes in both the parental species. Only pairs of linked loci having LOD scores higher than 10 were selected for this test. The results of the analysis showed that no interval in particular reflects significant inequalities in the frequency of recombination between the parents. Nevertheless, when all the intervals are included in the analysis by chi-square as a unit, the differences between parents become significant and in favor of the male C. eugenioides with 137 events of recombination compared with 100 events in the female C. liberica (X2 = 5.78, significant at α < 0.05, d.f. = 1).
Differential recombination between sexes has been studied in several species as a common phenomenon in nature, including species of agricultural interest (Burt et al., 1991; Plomion and O'Malley, 1996). In the apple tree, Malus pumila Mill., significant differences in recombination frequency have been detected between the parents of a mapping population, and when the difference in frequency are very high, it can have considerable affects on the determination of the correct order of loci (Maliepaard et al., 1998). It is possible that the probability of finding differences in recombination frequency between parents is greater in a cross between two individuals from different species, as it is found in this study. Nevertheless, in this work, the magnitude of those differences does not seem to considerably affect the individual intervals of the map distances. In this case the level of saturation of the map seems to play a very important role in decreasing the effects of differential recombination between the parents of the mapping population.
Conclusions
- The segregation analysis in the descendants of the interspecific cross between C. liberica and C. eugenioides in this study, and specially based on the results presented with respect to the amounts of each segregant type, it is possible to conclude that SSRs markers are a very informative approach for genetic analysis in coffee and appears that the majority of this type of loci are genetically very informative.
- The phenomenon of segregation distortion detected in this study is comparable with that found in other studies. Using a criterion of a ∼ 0.01, 40 loci were distorted, corresponding to 23.5%. These values are considered to be average for plants and, given the fact that the population under investigation was derived from an interspecific cross; it is very probable that this aspect may have played an important role in the segregation distortions found here.
- In addition, based on the proportions of segregant and distorted loci for each of the parents, it was evident that C. liberica is the parental species that presented the major proportion of segregant loci, and that C. eugenioides is the parental species with greater tendency to cause segregation distortions.
- In this study, a clear tendency to exhibit a major proportion of null alleles was detected in the paternal C. eugenioides. In some cases it was possible to verify the presence of null alleles by changing the PCR conditions or the sequence of primers. This allows the validation of the segregant types, thereby avoiding the ambiguity of genotype scoring for plant segregating analysis.
- Considering only the more informative loci (segregant types abxcd or efxeg), differences between parents with regard to recombination frequencies when all the intervals are included allows to suggest that, in this cross, C. eugenioides was the parental with major tendency to chromosomal recombination during meiosis.
Acknowledgments
We are grateful to Gabriel Cadena for his permanent support during the development of the project; to Gustavo Mendoza and Juan Carlos Montoya for technical support in laboratory procedures. This work was supported with funds from Federación Nacional de Cafeteros de Colombia, Cenicafé, and Ministerio de Agricultura y Desarrollo Rural (MADR).
References
Barre, P.; Louarn, J.; Charrier, A.; Hamon, S.; and Noirot M. 1997. Introgression of quality traits from Coffe pseudazanguebariae to C. liberica var Dewevrei. In: XVII Colloque Scientifique International Sur Café, Nairobi, Kenya. [ Links ]
Barret, B.; Griffiths, A.; Schreiber, M.; Ellison, N.; Mercer, C.; et al. 2004. A microsatellite map of white clover. Theor. Appl. Genet. 109:596 - 608. [ Links ]
Bridson, D. M. and Verdcourt, B. 1988. Flora of tropical East Africa - Rubiaceae Part 2. Balkema, Bookfield, Rotterdam. [ Links ]
Burt, A.; Bell, G. and Harvey, P. 1991. Sex differences in recombination. J. Ecol. Biol. 4:259 - 277. [ Links ]
Chen, X.; Cho, Y. G.; and McCouch, S. R. 2002. Sequence divergence of rice microsatellites in Oryza and other plant species. Mol. Genet. Genom. 268:331 - 343. [ Links ]
Eckard, N. A. 2001. A sense of self: The role of DNA sequence elimination in allopolyploidization. Plant Cell 13:1699 - 1704. [ Links ]
Grattapaglia, D. and Sederoff, R. 1994. Genetic linkage maps of Eucalyptus grandis and Eucalyptus europhylla using a pseudo-testcross: Mapping strategy and RAPD markers. Genetics 137:1121 - 1137. [ Links ]
Harushima, Y.; Kurata, N.; Yano, M.; Nagamura, Y.; Sasaki, T.; et al. 1996. Detection of segregation distortions in an indica-japonica rice cross using a high-resolution molecular map. Theor. Appl. Genet. 92:145 - 150. [ Links ]
International Coffee Organization. World Coffee Trade Section. Available at: http://www.ico.org/trade_e.asp?section=about_coffee [Reviewed in February 7th 2014] [ Links ]
Jiang, C. and Zeng. Z. B. 1997. Mapping quantitative trait loci with dominant and missing markers in various crosses from two inbred lines. Genetics 101:47 - 58. [ Links ]
Ky, C. L.; Louarn, J.; Guyot, B.; Charrier, A.; and Hamon, S.; et al. 1999. Relations between inheritance of chlorogenic acid contents in an interspecific cross between Coffea pseudozanguebariae an Coffea liberica var 'Dewevrei'. Theor. Appl. Genet. 98:628 - 637. [ Links ]
Ky, C.L.; Barre, P.; Lorieux, M.; Trouslot, P.; Akaffou, S.; et al. 2000. Interspecific genetic linkage map, segregation distortion and genetic conversion in coffee (Coffea sp.). Theor. Appl. Genet. 101:669 - 676. [ Links ]
Lashermes, P.; Combes, M. C.; Robert, J.; Trouslot, P.; D'Hont, A.; et al. 1999. Molecular characterization and origin of the Coffea arabica L. Genome. Mol. Gen. Genet. 261:259 - 266. [ Links ]
Lashermes, P.; Combes, M. C.; Prakash, N. S.; Trouslot, P.; Lorieux, M.; et al. 2001. Genetic linkage map of Coffea canephora: effect of segregation distortion and analysis of recombination rate in male and female meiosis. Genome 44:589 - 596. [ Links ]
Lashermes, P.; Combes, M. C.; Ansaldi, C.; Elijah Gichuru; and Noir, S. 2011. Analysis of alien introgression in coffee tree (Coffeaarabica L.). Mol. Breeding 27:223 - 232. [ Links ]
López, G. and Moncada, M. P. 2005. Construcción de un mapa de ligamiento genético preliminar de Coffea liberica x C. eugenioides. Revista del Centro Nacional de Investigaciones de Café - Cenicafé (ISSN 0120-0275). 35(4):319 - 338. [ Links ]
López, G. A.; McCouch S. R.; and Moncada, M. P. 2013. A genetic map of an interspecific diploid pseudo testcross population of coffee. Euphytica 192(2):305 - 323. [ Links ]
Luo, Z.W.; Hackett, C. A.; Bradshaw, J. E. ; McNicol, J. W.; and Milbourne, D. 2001. Constrtuction of a genetic linkage map in tetraploid species using molecular markers. Genetics 157:1369 - 1385. [ Links ]
Maliepaard, C.; Alston, F. H. ; Van Arkel, G.; Brown, L. M. ; Chevreau, E.; et al. 1998. Aligning male and female linkage maps of apple (Malus pumila Mill.) using multi-allelic markers. Theor. Appl. Genet. 97:60 - 73. [ Links ]
Maliepaard, C.; Jansen, J.; and Van-Ooijen, J. W. 1997. Linkage analysis in a full-sib family of an outbreeding plant species: overview and consequences for applications. Genet. Res. Cambr. 70:237 - 250. [ Links ]
McCouch, S. R.; Kochert, G.; Yu, Z. H.; Wang, Z. Y.; Khush, G. S.; et al. 1988. Molecular mapping of rice chromosomes. Theor. Appl. Genet. 76:815 - 829. [ Links ]
Moncada, P. and McCouch, S. R. 2004. Simple sequence repeat diversity in diploid and tetraploid Coffea species. Genome 47:501 - 509. [ Links ]
Montoya, J. C.; Moncada, M. P.; and López, G. A. 2006. Optimización y Caracterización de SSRs en una Muestra del Banco Germoplasma de Coffea de Cenicafé. Fitotecnia Colombiana (ISSN 0123- 1286). 6(2):44 - 51. [ Links ]
Paillard, M.; Lashermes, P.; and Pétiard, V. 1996. Construction of a molecular linkage map in coffee. Theor. Appl. Genet. 93:41 - 47. [ Links ]
Pearl, H. M.; Nagai, C.; Moore, P. H.; Steiger, D.L.; Osgood, R. V.; et al. 2004. Construction of a genetic map for arabica coffee. Theor. Appl. Genet. 108:829 - 835. [ Links ]
Plomion, C. and O'Malley, D. M. 1996. Recombination rate differences for pollen parents and seed parents in Pinus pinaster. Heredity 77:341 - 350. [ Links ]
Prakash N. S.; Marques, D. V.; Varzea V. M.; Silva, M. C.; Combes M. C.; et al. 2004. Introgression molecular analysis of a leaf rust resistance gene from Coffea liberica into C. arabica L. Theor. Appl. Genet. 109:1311 - 1317. [ Links ]
Ritter, E.; and Salamini, F. 1996. The calculation of recombination frequencies in crosses of allogamous plant species with applications to linkage mapping. Gen. Res. 67:55 - 65. [ Links ]
Romero, J. V. and Cortina-Guerrero, H. 2004. Fecundidad y ciclo de vida de Hypothenemus hampei Coleoptera: Curculionidae: Scolytinae en introducciones silvestres de café. Rev. Cenicafé 553:221 - 231. [ Links ]
Van der Vossen, H. A. 1985. Coffee selection and breeding. In: Coffee. botany, biochemistry and production of beans and beverage. Edited by M.N. Clifford and K.C. Willson. Avi Publishing Company, Westport, Conn. p. 48 - 96. [ Links ]
Van-Ooijen, J. W.; and Voorrips, R. E. 2001. Join- Map® 3.0, Software for the calculation of genetic linkage maps. Plant Research International, Wageningen, Holanda. [ Links ]
Xu, Y.; Zhu, L.; Xiao, J.; Huang, N.; and McCouch, S. R. 1997. Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid and recombinant inbred populations in rice (Oryza sativa L.). Mol. Gen. Genet. 253:535 - 545. [ Links ]
Segregation analysis of molecular markers in a population derived from Coffea liberica Hiern x C. eugenioides L. [ Links ]