Introduction
Cocoa, Theobroma cacao L., is the most economically important species in the Malvaceae family. In its natural habitat, cocoa grows in the understory of evergreen tropical rainforest. It often grows in clumps along river banks, where the roots may be flooded for long periods of the year. Cocoa grows in low elevations, usually below 300 meters above sea level, in areas with 1000 to 3000 mm rainfall per year. It was domesticated in the Amazon basin and today it is widely cultivated on roughly 17,000,000 acres (27,000 sq mi; 69,000 km2) worldwide, principally, in West Africa, Central and South America and Southeast Asia (Almeida & Valle, 2009).
The fruit is an egg-shaped red to brown berry (commonly referred to as a 'cocoa pod'), 15 to 25 cm long, with a more or less knobby surface and lines from top to bottom. The pod contains 30 to 40 seeds, each of which is surrounded by a bitter-sweet white pulp. In the wild, the seeds are dispersed and they serve as food for different mammals like agoutis and monkeys. When the seeds are dried and fermented in the sun they get brownish red, and become cocoa beans. Their cultivation is primarily intended for providing cocoa almonds used in the production of chocolate and other derivatives, such as jellies, ice creams, and juices (Almeida & Valle, 2009), as well as by products that can be processed in pharmaceutical and cosmetic industry (Adeagbo et al., 2008; Karim et al., 2014).
Around the world, more than five million cocoa farmers depend on this product for their livelihood, according to the World Cocoa Foundation, which puts annual cocoa production worldwide at 3.7 million tons, valued at $11.8 billion. Biotic stress, expressed in fungal diseases and insect attacks (Argout et al., 2008, Purdy & Schimidt, 1996), and such abiotic factors as irradiance, droughts and floods (Sena & Kozlowski, 1986) can produce significant production losses.
The occurrence of pests and diseases to which cocoa is subject, along with climate change, highlight the need for new varieties able to respond to these threats. Breeders rely on the genetic diversity conserved in field germplasm collection to create new varieties, because cocoa recalcitrant seeds cannot be stored in a conventional germplasm collection. In an effort to improve the genetic diversity available to breeders, and to ensure the future of field germplasm collection, experts have drawn up a global strategy for conservation and use of cocoa genetic resources, namely, the Foundation for a Sustainable Cocoa Economy. The strategy has been adopted by cocoa producers and their clients. It seeks to improve characterization of cocoa diversity, sustainability, diversity, and usefulness of cocoa collections, and to ease the access to a better information about the conserved material.
In Ecuador, cocoa is one of the main tropical industrial crops and there is a great interest in promoting field culture for elite varieties to improve national competitiveness as well as productivity toward the industrial sectors. Major biotic threats causing important crop yield losses affect the pod, which constitutes an interesting study material for discovering pathogen resistance genes and proteins. The recent sequencing of cocoa genome (Argout et al., 2011), and the deployment of both nucleotide and protein sequences in public databases, has opened new perspectives in research, such as cocoa high-coverage proteomic analysis. Genome-wide sequences allow for the assignment of mass spectrometry (MS) data to particular gene products, even distinguishing between paralogs, thus leading to a confident identification of cocoa proteins, that is, to determine the exact genes involved and not just molecular functions of differential proteins (Sellés-Marchart et al., 2008).
Efficient methods of protein extraction are essential to successfully apply proteomic analyses in plants and in particularly important agronomic crops, such as cocoa. Standard protocols have been proposed for various types of samples, but the particularities of many samples require the use of specific protocols optimized according to the objective of the study, the specific type of tissue, and the age of the organ (Görg et al., 2004; Islam et al., 2004). T. cacao, in particular, has a very high level of interfering substances, such as polysaccharides and phenolic compounds (Gesteira et al., 2003), that can prevent the isolation of suitable protein and that possibly explain the absence of data in the literature about proteome analysis of T. cacao pods.
In order to obtain quality proteins and successfully apply proteomic analyses, it is necessary to develop an efficient protocol for protein extraction specifically from pods of this species. Thus, we tested and compared two protein extraction methods reported in literature to have been successful with other recalcitrant tissue types. Concretely, we evaluated two methods (A and B) for the preparation of tissue for total protein extraction using phenol/SDS extraction protocol.
Material and methods
Plant materials
Cocoa pods (Theobroma cacao L.) were harvested at optimal ripeness from trees cropped in the experimental orchards of La Represa at Quevedo, Universidad Tecnica Estatal de Quevedo-Ecuador (UTEQ), Ecuador (79º 30´ 23´´ W, 01º 00´ 35´´ S, 90 m. a. s. l.). After cocoa seeds were removed pods were cut into small pieces (1x1 cm); batches of 50 g of tissue were liquefied in 1 % ascorbic acid (w/w), then grounded in liquid nitrogen using a mortar and pestle and stored at -80 ºC in an ultrafreezer until use.
Sample tissue preparation
Two protocols were tested for the preparation of pod tissue for total protein isolation, one as described by Wang et al., 2003 with some modifications (Method A) and the other as described by Sellés et al., 2008 with some modifications (Method B) both illustrated below (Figure 1).
Method A: about 0.1 - 0.3 g of fine tissue powder was placed in 1.5 mL microtubes and resuspended in 1mL cold 20 % trichloroacetic acid (TCA) in acetone. After vortexing thoroughly for 30 s, the tubes were centrifuged at 10000 x g for 5 min (4 ºC). The supernatant was discarded, and the pellet was washed 3 - 4 times or until the supernatant became colorless, then with cold 20 % trichloroacetic acid (TCA) in water twice, and finally with cold 80 % acetone twice. In each wash, the sample was centrifuged at 10000 x g for 5 min (4 ºC). The final pellet was dried at room temperature and used either for protein extraction or stored at -20 ºC for future use.
Method B: 1 g of the fine tissue powder was homogenized in 10 mL of cold 0.1 M sodium phosphate pH 7.0 buffer containing 20 mM ascorbic acid. The homogenate was filtered through 8 layers of gauze, constituting the crude extract. It is used either for protein extraction or stored at -20 ºC for future use.
Protein extraction
The protein extract was prepared by phenol/SDS method (Wang et al., 2003) with some modifications. Briefly, the cleaned pod dry powder (about 0.1 - 0.3 g) or the crude extract (0.4 mL) was supplemented with 0.8 mL Tris-saturated phenol pH 8.0 (Sigma St. Louis, MO, USA) and 0.8 mL SDS buffer (30 % sucrose, 2 % SDS, 0.1 M Tris-HCl, pH 8.0, 5 % 2-mercaptoethanol) in a 2.0 mL microtube. The mixture was vortexed thoroughly for 30 s and incubated with orbital shaking on ice for 1 h. The phenol phase was separated by centrifugation at 10000 x g for 20 min at 4 ºC. The upper phenol phase was recovered and pipetted to fresh microtubes (0.2 mL for 1.5 mL tube, 0.4 mL for 2.0 mL tube). The remaining aqueous phase was re-extracted with 0.8 mL Tris-saturated phenol and 0.8 mL SDS buffer. After phase separation, white SDS complex often appears at the interphase. Care should be taken not to disturb the interphase by pipetting. Proteins were precipitated from the pooled phenol phases by adding 5 volumes cold 0.1 M ammonium acetate in methanol, incubating at -20 ºC overnight and collecting by centrifugation at 10000 x g for 10 min at 4 ºC. The protein pellet was washed twice with 0.1 M ammonium acetate in methanol and twice with chilled 80 % acetone, and then finally let to dry at room temperature. The protein concentration of samples solubilized in SDS-PAGE sample buffer (0.5 M Tris-HCl pH 6.8, 10 % SDS, 10 % Glycerol, 14.3 M β-mercaptoethanol and 1 % Bromophonol blue) was determined by Bradford method (Bradford, 1976) with bovine serum albumin as a standard over three replicated assays for each sample diluted four, five and six times in distilled water.
Electrophoresis
SDS-PAGE was performed according to Laemmli et al., 1970 in Bio-Rad Mini-Protean II system (7 cm × 10 cm minigels). Fifteen microliters of 1 µg.µL-1 total protein extract were boiled for 5 min and loaded per well. Proteins were resolved in 12.5 % polyacrylamide gel and visualized after MS-compatible silver staining (Shevchenko et al., 1996). Briefly, after electrophoresis, the gel slab was fixed in 50 % methanol, 5 % acetic acid in water for 20 min. It was then washed for 10 min with 50 % methanol in water and additionally for 10 min with water to remove the remaining acid. The gel was sensitized by 1 min incubation in 0.02 % sodium thiosulfate, and it was then rinsed with two changes of distilled water for 1 min each. After rinsing, the gel was submerged in chilled 0.1 % silver nitrate solution and incubated for 20 min at 4 °C. After incubation, silver nitrate was discarded, and the gel slab was rinsed twice with water for 1 min and then developed in 0.04 % formalin [35% formaldehyde in water] in 2 % sodium carbonate with intensive shaking. After the developer turned yellow, it was discarded and replaced with a fresh portion. It is essential that the developing is carried out in an absolutely transparent solution. After the desired staining intensity was achieved, the development was terminated by discarding the reagent, followed by washing of the gel slab with 5 % acetic acid. Developed gels were completely transparent when the sensitization step with sodium thiosulfate was included. Silver-stained gels were stored in a solution of 1 % acetic acid at 4 °C.
Results and discussion
Sample preparation is one of the most crucial, yet problematic, steps to yield high-quality proteins. T. cacao has a very high level of interfering substances, such as polysaccharides and phenolic compounds (Gesteira et al., 2003), that could prevent the isolation of suitable protein and possibly explain the absence of data in the literature about proteome analysis of T. cacao pods. We evaluated and compared two methods (A and B) of tissue preparation for total protein extraction using phenol/SDS extraction protocol (Wang et al., 2003). The difference between the two methods compared in this study was the tissue handling before protein phenol-based extraction: in method A an extensive washing of pod tissue dry powder was made, whereas in method B an aqueous buffer crude protein extract was prepared. With Method A, 1.0 g lyophilized pods of T. cacao typically yielded approximately 1.5 mg protein, which was more than the 1 mg yield obtained by Method B. The quality and amount of protein extracted using the two methods (A and B) was monitored by one-dimensional SDS-PAGE (Figure 2). Both methods proved to be efficient for the extraction of cocoa pod proteins. In 1-DE gel, proteins isolated by Method A, showed about 20 sharp polypeptide bands ranging from 14-55 kDa (Figure 2A). Proteins isolated by Method B showed about 17 sharp bands of peptides ranging from 14-166 kDa (Figure 2B). Highly reproducible 1-D gel-banding profiles were observed using Method A. In contrast, protein extraction using Method B failed to show reproducible 1-D gel-banding patterns; numerous major bands were present in one technical replicate and absent from the other (data not shown), suggesting that Method B may need to be refined further for the extraction of cocoa pod proteins. Both experiments were repeated at least three times to confirm the reproducibility.
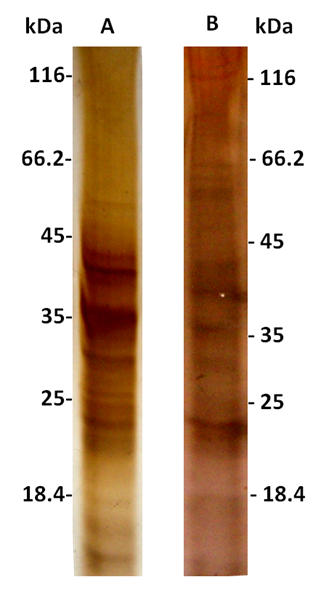
Figure 2 SDS-PAGE electrophoresis of cocoa pod protein extracts. A) Method A of total protein extraction. B) Method B of total protein extraction. Fifteen micrograms total protein were loaded per lane on 12.5% polyacrylamide gel and visualized with silver. PageRuler Unstained Protein Ladder was used as reference.
In plants, glycosylations is mostly encountered on secreted proteins, although some forms of glycosylation can also be found on several cytosolic or nuclear proteins (Fitchette et al., 2007). The variation among 1-D gels by Method B could be due to different glycosylation state of proteins, as it was reported in plants by Saravanan et al. (2004). 1-D gels were not treated with a glycoprotein-specific stain to determine if there was a bias in the glycosylation state of proteins. Therefore, future study could further define differences between the extractions.
The differences in the proteomes isolated using the two extraction types also raise the question of whether one method of protein extraction is sufficient to scan proteomes. One idea might be to use a tandem extraction protocol. Subfractionation schemes have been used previously to achieve greater proteome coverage. Alternatively, proteins obtained through different extraction procedures might be combined and analyzed simultaneously. With either suggested approach, it would be necessary to ensure statistically that each extraction step was reproducible.
Quantitative approaches require minimal variation between replicates so one can attribute any change in protein expression to treatment conditions (Karp et al., 2008). Simply having a method that extracts large quantities of protein is not sufficient unless it is reproducible. Therefore, the Method B used here may not be suitable for a gel-based quantitative-comparative approach. Rather, it may be more suited to applications where the primary goal is protein discovery rather than correlating variation between samples to a biological activity. However, the Method A showed reproducible thus demonstrating that Method A extraction method is a reliable method for preparing cocoa pods proteins when a gel-based quantitative-comparative approach is the target.
Conclusion
In conclusion, through Method A, we succeeded in isolating high-quality proteins from T. cacao pods. The 1-DE gels obtained were of high quality and could be used for protein identification by MS. It is expected that our protocol be also applied for other recalcitrant plant pods. The differences in the proteomes isolated using the two extraction types also raise the question of whether one method of protein extraction is sufficient to scan proteomes. Therefore, a combination of extraction approaches is recommended for increasing proteome coverage when using gel-based separation techniques.