Introduction
Lulo (Solanum quitoense Lam.) belongs to the Solanum genus and the Lasiocarpa section, which includes about 12 species (Bohs, 2004). In Latin America, economically profitable crops belong to the species Solanum quitoense and Solanum sessiliflorum (Heiser, 1985). In Colombia, S. quitoense, has gained importance in the agroindustrial sector for juices, yogurt, flavoring, soft drinks and processed foods because of its appearance, bittersweet flavor, and fragrance, ease of cultivation, continuous production, and high yields (Lobo et al., 2007).
However, this species is still in domestication process and have some important traits as follows: allogamy, narrow ecological adaptation, presence of thorns on stems and leaves, anthocyanins in different organs, trichomes, latency and high number of seeds per berry, andromonoecy, rapid oxidation of the juice and presence of ideoblasts in the leaves, as seen in individuals in the weed-wild complex (Lobo, 2006).
In Colombia, three systematic collection processes have been conducted for S. quitoense, and related taxa, covering the departments of Antioquia, Boyaca, Caldas, Cauca, Huila, Magdalena, Nariño, Norte de Santander, Putumayo, Quindio, Santander, Tolima and Valle del Cauca (Lobo et al., 2007). The potential use of a collection depends on the materials knowledge. In this country, studies conducted for characterization and evaluation of S. quitoense collections and related species, have allowed exhibiting the existence of polymorphism (Fory et al., 2010; Lobo et al., 2007; Riascos et al., 2012). Similarly, molecular studies have revealed significant polymorphism (Fory et al., 2010), and a study conducted with AFLP markers (Fory et al., 2010), found greater genetic variability in interspecific hybrids, S. hirtum × S. quitoense, as compared to the parentals, which points out the potential for increasing the genetic basis of S. quitoense for breeding programs.
The advance of fruit genetic breeding programs by conventional procedures is difficult and expensive, mainly due to the very long growing seasons, inter- and intraspecific incompatibility, presence of important attributes in wild species and high degree of heterozygosity (Lobo, 2006). The use of biotechnological tools in these programs would improve the selection process, facilitating control and management of the genetic basis of populations, saving resources and ensuring genetic purity of new varieties (Moose & Mumm, 2008). Therefore, it is necessary to implement strategies for the evaluation and characterization of S. quitoense materials with farmers in different regions of the country.
Among the known molecular markers, RAM (Random Amplified Microsatellites) are very useful for measuring genetic diversity in plants and animals to detect differences among families, species and within species. In addition, Muñoz et al. (2008), showed that the variation basis of individuals, have allowed the selection of specific regions within the DNA molecule for determined studies, the number of detectable polymorphisms is theoretically unlimited and allows for the analysis of information that is both expressed and non-expressed. This method is feasible to implement in small laboratories in terms of equipment and facilities cost; it requires no prior sequencing knowledge or the use of radioactive isotopes; therefore, can be used in characterization studies of genetic diversity (Morillo et al., 2014).
In order to establish a strategy and plant management for phytogenetic resources for S. quitoense in the department of Boyaca- Colombia especially in the Province of Neira, it is necessary to start studies on morphological, agronomic, physiological and molecular characterizations to know the genetic diversity and generate basic necessary information to obtain sustainable solutions for the problems of low levels of technology in production, common in S. quitoense cultivation, such as lack of homogeneity in organoleptic characteristics in materials used in beverages and jams, genetic transformation, which allows for a shorter growth cycle for production in less time and tolerance to pests and diseases throughout plant breeding program (Lobo et al., 2007). Within this context, this research aimed to establish a molecular characterization of lulo (Solanum quitoense Lam.) materials in the Province of Neira, Boyaca-Colombia using random amplified microsatellite markers (RAM).
Materials and methods
Biological material
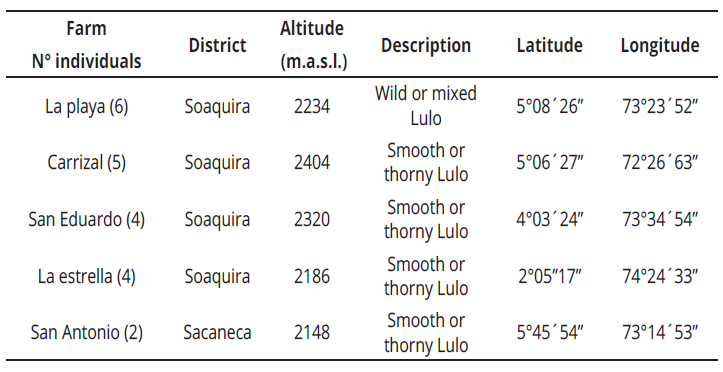
Young leaves were collected from a total of 21 S. quitoense materials in the province of Neira, municipality of Pachavita in the Soaquira district, department of Boyaca, Colombia; located at an altitude of 1985 m. a. s. l. with an average temperature of 16°C and in the Sacaneca district, at an altitude of 2148 m. a. s. l., with an average temperature of 17°C (Table1).
Molecular characterization
Molecular characterization was carried out at the Universidad Pedagógica y Tecnológica de Colombia. Boyacá-Tunja, Colombia. DNA extraction was according to Dellaporta et al. (1983).
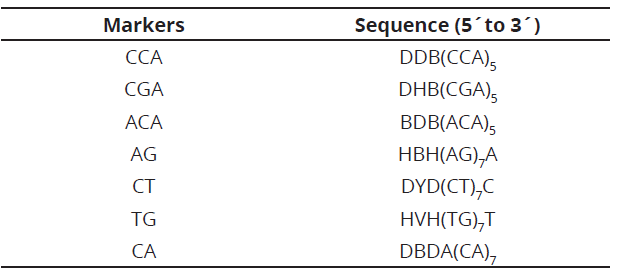
Genomic DNA of displayed samples was in 0.8% agarose gels, stained with z-vision in a MaxiCell Primo EC-340 electrophoresis gel system chamber. A Hoefer Dyna Quant 200 fluorometer, was used to determine the concentration of each genotype and HPLC water, was used for the dilution (10 ng μL-1), at a total volume of 100 µL and stored at -20°C. Seven markers, synthesized by Technologies Inc. (Table 2), were used for RAM analysis.
For RAM amplification, a reaction cocktail was prepared in a sterile microcentrifuge tube (1.5 mL) at a final volume of 25 µL. The reaction mixture was prepared with 1X buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 1U DNA Taq polymerase, 2 uM of marker primer and 10 ng of genomic DNA.
The amplification was carried out in a PTC 100 Programmable Thermal Controller thermocycler (MJ. Research, Inc.). Initial denaturation was at 95 ° C for 5 min; denaturation at 95 ° C for 30 sec, annealing at a temperature of 50 ° C (primer AG CA), 55 ° C (primer CCA-TG-CT) and 58 ° C (primer CGA) for 45 sec, one final extension of 72 ° C for 2 min, 37 cycles from denaturation to extension, and a extension at 72 ° C for 7 min. Products were separated by electrophoresis in 1.2% high resolution agarose gel at 90 volts for 3 h, displayed in an ultraviolet light transilluminator.
Statistic analysis
A binary matrix was created, absence (zero) and presence (one). The genetic similarity among individuals was calculated using the Nei-Li similarity coefficient. The grouping analysis was carried out with UPGMA method and a dendrogram was created with the NTSYS statistical package (Numerical Taxonomy System for personal Computer, version 2.02 PC ™). The unbiased heterozygosity and polymorphic loci percentage were estimated to evaluate the genetic diversity using the TFPGA (Tools for Population Genetic Analyses) statistical package, version 1.3. ®.
Results and discussion
The analysis with the Nei-Li coefficient at a 0.60 level of similarity differentiated in S. quitoense materials into three groups (Figure 1). Group I, had three S. quitoense materials from the Soaquira district-Boyaca, Colombia in the following farms: La Playa, San Eduardo and Carrizal, which were characterized as wild S. quitoense material, conserved by farmers in that area, exhibiting traits of incipient domestication.
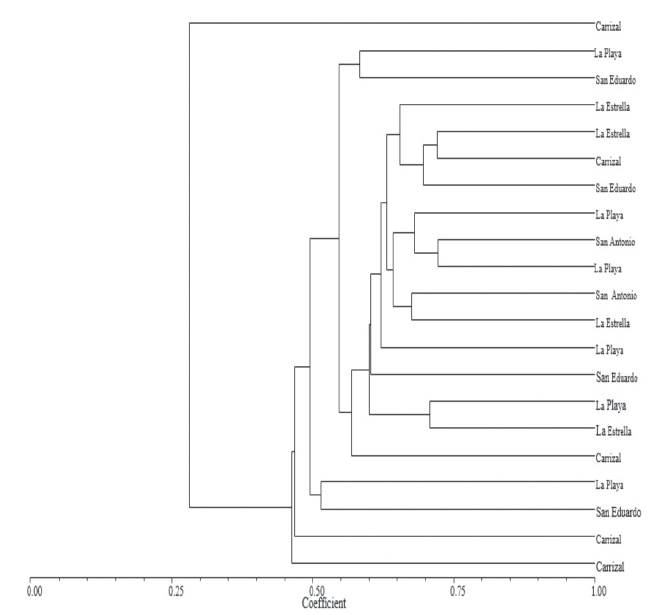
Figure 1 Dendrogram of 21 lulo materials (S. quitoense Lam.) based on the coefficient of Nei and Li.
This group contained material from the Carrizal farm, which had the lowest similarity value (0.25) with respect to rest of S. quitoense materials. This have allowed establishing specific morphological traits of S. quitoense materials and geographical separation, because these materials were collected at higher altitudes.
Group II, had S. quitoense materials collected in two districts, Soaquira and Sacaneca- Boyaca, Colombia demonstrating that gene flow may exist among these materials, throughout seed exchanges by farmers in this area. These S. quitoense materials were smooth (thornless) or thorny, thick and mixed, characterized by high coefficients of variation in the qualitative and quantitative characteristics. Traits, which are favorable for marketing, helping to solve the problem of reduced fruit size and weight in this country (Riascos et al., 2012).
Group III, with a level of similarity of 0.50, contained three S. quitoense materials, which were collected at the Carrizal farm, which had the highest genetic distance (0.45) from the other evaluated materials, which corroborates the fact that the materials of this farm, are highly variable and wild, maintained by this farmer and found at a higher altitude than the others.
In general, the groupings had a loose distribution of S. quitoense materials, with no relationship with the districts, but showed an exchange or gene flow among them. Furthermore, there were no associations for the presence or absence of thorns, as has been reported in other studies on S. quitoense genetic diversity (Riascos et al., 2012). The obtained results in this research, are different from those obtained by Fory et al. (2010), who reported low variability in cultivated species of S. quitoense, using AFLP markers. This difference is associated with the type of marker as follows: AFLP and RAM markers, are dominant and COS II markers are co-dominant, and are designed based on conserved exon in order to amplify intron sequences, which increases the possibility to find polymorphism.
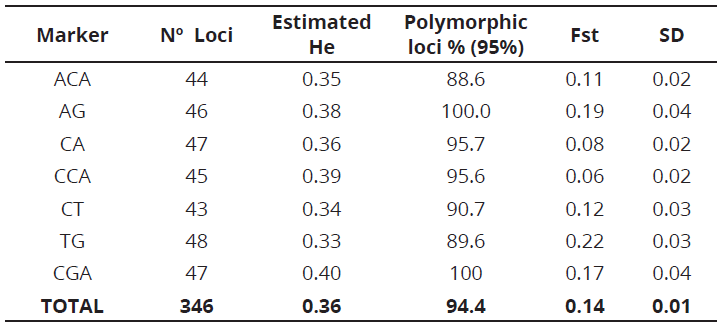
The seven RAM markers used in this study generated 346 alleles, which ranged from 43 for the CT marker to 48 for TG marker, with molecular weights between 300 and 1000 bp (Table 3). The number of bands was suitable for estimating parameters of genetic diversity when compared with the obtained results in other studies on S. quitoense and other Solanaceae species (Saliba-Colombani et al., 2000; Toquica et al., 2003; Fory et al., 2010). TG marker, performed the greatest contribution to the observed variation with a 0.22 Fst, which means it can be useful for other studies on the genetic diversity in the Solanum genus.
The average estimated heterozygosity values ranged from 0.33 for TG marker to 0.40 for CGA marker. The average value for all evaluated S. quitoense materials, was 0.36. In addition, the fact that S. quitoense is an allogamic species, but can also experience self-pollination and be an andromonoecious species must be taken into account (Enciso-Rodríguez et al. 2010). Ge et al. (2013), when analyzing the genetic diversity and structure of eggplant populations (Solanum melongena L.) in China using simple sequence repeat markers, found an expected heterozygosity value of 0.32, lower than the value reported in this study.
Other genetic diversity studies with codominant microsatellite markers on different species (S. elaeagnifolium, S. rostratum and S. tuberosum spp. tuberosum) of the Solanaceae family, have shown higher heterozygosity values, demonstrating the high variability present in these species and the potential of these markers to determine the genetic structure and diversity in natural populations (Vallejo-Marín et al., 2011; Zhu et al., 2013).
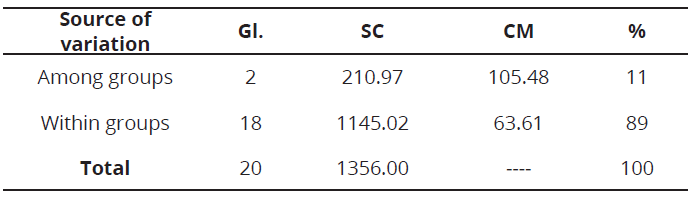
The coefficient of genetic differentiation (Fst = 0.14 ± 0.01), suggests an intermediate level of genetic differentiation, which may be due to the state of domestication of the evaluated materials and may also indicate that there is a moderate relationship between the geographical location and genetic group (Morillo et al., 2014). The molecular variance analysis AMOVA, showed the genetic variation observed in the evaluated S. quitoense materials, was mainly within groups, with 89 % (Table 4). This high variation could indicate the presence of higher levels of subdivision and hierarchy. The remaining 11% was due to the component of genetic variance among groups, which was significant (P≤0.001). Such genetic variation among groups, might be used for conservation and breeding of this species. Similar results have been reported in other studies of genetic diversity in the genus Solanum using microsatellite markers (De Galarreta et al., 2007; Mercati et al., 2015; Zhou et al., 2015).
Results of this study are consistent with Lobo (2006), who found greater variation at the subgroup or subpopulation level, which is correlated to the geographical distribution of S. quitoense in this country, because Colombia is a center of origin and has diverse agro-ecological conditions, which are appropriate for this species. Geographical barriers, adaptation to different ecological niches, the reproduction mode of the species, and type of molecular markers used, could influence results (Lobo et al., 2007).
In general, high genetic variability was observed in S. quitoense materials, both cultivated and wild, contrary to what was reported by Fory et al. (2010), who found a reduced genetic variability in cultivated species of S. quitoense and species related to the Lasiocarpa section of the Colombian collection, which was mainly attributed to the founder effect. Owing to the low degree of domestication, is common that exist a high heterozygosity in Andean fruits, due to a mechanism of adaptation and survival to changing environmental conditions. However, S. quitoense is a primitive plant with a low degree of domestication, and stability and adaptability, which are restricted to very specific niches with direct natural variation and evolutionary processes (Lobo, 2006). Knowledge on the producers and communities has enabled the development and maintenance of local and primitive varieties (or so-called folk varieties), which have been the basis for the development of most crops, and are continuously subjected to selective pressures imposed by the spatiotemporal dynamics of growth and human being selection processes (Lobo, 2006).
The use of molecular markers in the evaluation and characterization of wild germplasm, provides great opportunity for a more precise determination of the relationships and genetic structure and evolution of the populations. In addition, it is an important tool for gene transfer assisted by markers to identify variants at the genome level. The introgression of interest traits throughout interspecific hybridization and conservation of wild materials is an alternative, which can help to identify elite materials that are adapted to local conditions and meet the needs of farmers, producers and consumers.
Conclusion
Lulo (Solanum quitoense Lam.) materials were grouped into three groups, which did not present any associations strictly related to the geographic area or thorn presence or absence. This research study revealed the existence of genetic variability in the evaluated S. quitoense materials in the Province of Neira-Boyaca, Colombia, which are readily available to be used in breeding programs aimed to find elite materials in this area.