INTRODUCTION
Mycoplasma spp. are a group of simple pleomorphic bacteria (around 1000 kbp), of the mollicute class, characterized by the lack ofa cell wall (Tenk 2005). Mycoplasma spp. can be located in several body sites in cattle, and, according to the site, it can produce different diseases like arthritis, otitis, keratoconjunctivitis, pneumonia, reproductive disorders and mastitis (González and Wilson 2003).
As a mastitis causing pathogen, Mycoplasma spp. are classified as highly contagious pathogens that can cause a severe inflammatory reaction in the udder, with a subsequent increase in somatic cell count (SCC) and/or severe clinical mastitis (Lysnyansky et al. 2016; Nicholas et al. 2016). Commonly, it responds poorly to antimicrobial treatments and it is usually unsuccessful as a result of the high antibiotic resistance of the mycoplasmas (Barberio et al. 2016). There is no effective therapy reported for mastitis, and the antibiotic treatment is, therefore, not considered in the treatment of the disease. The protocol described to control mastitis due to mycoplasma is to identify and segregate infected cows (Bushnell 1984; González and Wilson 2003; Aebi et al 2015).
It is described that the transmission of mycoplasma mastitis has different epidemiology and a different set of risk factors than other contagious mastitis pathogens, e.g. herd size (Fox et al. 2003). The frequency of isolation of Mycoplasma spp. is low compared to other pathogens in milk samples. However, this pathogen is reported almost everywhere, except for a few places around the globe (Arcangioli et al. 2011), with a prevalence close to 5% in the bulk tank milk in North American and European Union countries (Fox 2012; Nicholas et al. 2016). Mycoplasma mastitis appeared also to be present in Latin American countries such as Mexico, Brazil, Chile, Argentina, and Colombia (Boyacá), mostly with a low prevalence or as a mastitis outbreak in single dairy farms (Infante-Martínez et al. 1999; Ulloa 2013; Andrade-Becerra et al. 2014; Tamiozzo et al. 2014).
Bacteriological culture for Mycoplasma spp. requires special methods, such as adapted growing culture media, incubation temperature, CO2 concentration during incubation, and a prolonged time of incubation (3 to 10d), which can explain why this pathogen is probably under-reported (Fox 2012). In this regard, molecular diagnosis via highly specific and sensitive PCR methods is employed since year 2000, and allows for species identification and differentiation between mycoplasma and non-pathogenic bacteria occasionally found in milk, like Acholeplasma spp. (Pinnow et al. 2001; Boonyayatra et al. 2012a; Boonyayatra et al. 2012b).
Due to the presence of the pathogen in Colombia and the special requirements for the microbiological diagnosis, the objective of the current study was to estimate the prevalence of Mycoplasma spp. in bulk tank milk in Colombian dairy herds located in the mid-western region using microbiological and molecular diagnosis.
MATERIALS AND METHODS
Study design and sample size calculation
A longitudinal study with commercial dairy herds located in the mid-western region of Colombia located at variable altitudes ranging between 860 and 3700 m.a.s.l. was performed. Herds were affiliated to one of the four main dairy companies of the region. The planned sample size was 160 farms (Dohoo et al. 2009). It was estimated using a significance level of 95% (a =0.05), an estimated error of 1.8% and an a priori estimated herd level prevalence of1.4%. The latter was based on the prevalence reports of Mycoplasma spp. in bulk tank milk from São Paulo, Brazil (Manzi et al. 2018). Finally, a sampling frame of 303 herds was created from the four dairy companies; a total of220 dairy farms were randomly selected and included in the study. Of these 220 farms, 160 were randomly selected to determine the prevalence of Mycoplasma spp. based on microbiological and molecular analysis of bulk milk samples. On the remaining 60 herds, bulk milk was tested for the presence of Mycoplasma spp. via PCR only. Finally, 83 farms were not included in the study, but their SCC data was used to explore selection bias.
Farms selection and sampling
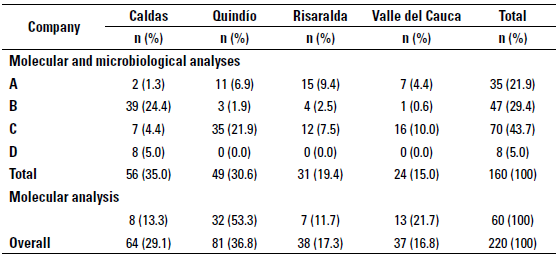
Levels of the stratified random sampling used to select 160 farms out of the 220 farms included in the study for microbiological and molecular analyses were determined by dairy company and geographical location, provinces included were: Caldas (n = 56), Quindío (n = 49), Risaralda (n = 31), and finally Valle del Cauca (Northern region) (n = 24) (table 1). The remaining 60 farms included for molecular analysis only were located as shown in table 1. The research Inclusion criteria for the farms were: to area corresponds to 38 municipalities form have a bulk tank and to deliver the milk the provinces mentioned before (figure 1). up to 2d after milking to one of the four dairy companies selected for the study. However, bulk tanks which included milk from more than one producer or farms with so-called "communitarian bulk tanks" and bulk tanks without refrigeration temperature were excluded from the study.
TABLE 1 Distribution of herds classified by geographical location and dairy company for milk delivery
Source: self-made.
Source: self-made.
FIGURE 1 Geographical distribution of the provinces and municipalities included in the study area.
A sterile sample of bulk tank milk was monthly collected for up to three samplings per farm between April and July of 2018. Before the sample was collected, the milk was agitated for 5-10 min and then, using a clean sanitized dipper, a total volume of 100 mL of milk was divided into two aliquots; the first one was combined with an 8mg bronopol tablet (Broad Spectrum Microtabs II™, Advanced Instruments) and was used for determination of the bulk milk SCC by flux cytometry (CombiFossTM, Foss), and the second one did not contain bronopol and was used for the detection of Mycoplasma spp. Samples were transported to the laboratory (Universidad de Caldas, Manizales, Colombia, for molecular and microbiological analyses and Universidad de Antioquia, Medellín, Colombia, for determination of SCC) under refrigeration temperature (4° C). During the first sampling, all the farms fulfilling the inclusion criteria in the region were sampled in order to allow for comparison between the farms included in the study and those which were eventually excluded from the study.
Microbiological analyses
Samples for microbiological analyses were processed within the first 24h after sampling, due to the detrimental effects of long periods of storage or freezing in the bacteria (Punyapornwithaya et al. 2009). For the microbiological analyses of bulk tank milk, 100 µL of milk was inoculated onto 65x15 mm petri dishes with Mycoplasma agar medium (Mycoplasma agar base CM401 and Mycoplasma supplement-G SR0059; Oxoid) supplemented with 67pg/ mL of cefoperazone (Cefoperazone sodium salt C4292, Sigma-Aldrich) (Arcangioli et al., 2011), no pre-enrichment steps were done.
Plates were incubated at 37° C with a humidity of 10% of CO2 during 10d, searching for suspicious bacterial fried egg colonies at days 3, 5, 7 and 10 with a stereoscopic microscope magnification of 40x (Hogan et al. 1999). Suspicious colonies were processed with the aim of obtaining pure colonies.
The purification of the colonies was made by two methods as follows: the suspicious colony was inoculated into 4 mL of mycoplasma broth (Mycoplasma Broth base CM402, and Mycoplasma supplement-G SR0059; Oxoid), in case that the colony was attached to agar medium, the last was cut using a sterile loop. Then, the piece of agar was introduced into the mycoplasma broth. Afterwards, the sample was homogenized with a vortex at low speed, and finally 10 µL of the broth was cultured in an agar. Both the agar and broth were incubated during 48h-72h until growing of bacteria was observed. The pure colonies were collected from the agar plate with a loop and resuspended into 1 ml of ultrapure water for genomic DNA extraction and 1-3 colonies were placed in 10 of ultrapure water with a straight loop for microwave-based method DNA extraction. The broth was cryopreserved at -84° C for further use, adding 30% (v/v) of glycerol (Boonyayatra et al. 2012b).
Genomic DNA extraction for PCR
Genomic DNA was extracted from bacterial cultures and directly from the milk. Genomic DNA extraction began with 1 mL of ultrapure water with suspended bacteria, those were centrifuged at 10,000g for 2 min, the supernatant was discarded, and the centrifugation step was repeated. Then, a commercial spin column-based method (Bacteria DNA Preparation Kit, Jena Bioscience) was used following the manufacturer's recommendations.
Microwave based method genomic DNA extraction was employed placing the tube with 1-3 colonies in a microwave oven during 2 minutes, then it was used as DNA template in the PCR without any additional extraction methods (Cobo-Ángel et al. 2017).
Genomic DNA extraction from the milk was done using an aliquot of 1 mL of milk, centrifuged at 10,000 g for 10 minutes, the fatty layer was removed using a sterile cotton swab, followed by a 2 min centrifugation at the same speed. After this, the supernatant was discarded, the pellet was resuspended in 500 µL of sterile saline solution followed by a 2 min centrifugation at 10.000 g, finally the supernatant was discarded, and the DNA extraction process began with the same commercial kit mentioned before (Cremonesi et al. 2006; McDonald et al. 2009; Volk et al. 2014).
Quality and quantity of DNA were assessed using a fluorospectometer (Nanodrop™ 1000, Thermo Fisher Scientific Inc, Waltham, MA, USA). Samples with > 10 ng/µL were used for molecular analysis, and those over 50 ng/µL were diluted with ultrapure water to 30 ng/µL.
Molecular identification
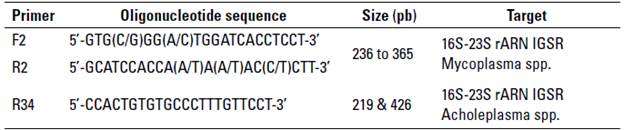
The PCR reaction followed the methodology suggested by (Boonyayatra et al. 2012b), with a final volume reaction of 50 µL, using 25 µL ofa commercial master mix (MangoMix, Bioline), 5 µL of DNA template (10 ng/[L), 1 [L of the primers shown in table 2, and 17 µL of ultrapure water. The PCR program was done as follows: initial denaturation at 94° C for 30 s, followed by 35 cycles ofdenaturation at 94 °C for 30 s, primer annealing at 55 °C for 2 min, and extension at 72 °C for 2 min. A final extension performed at 72 °C for 5 min. The final PCR products were electrophoresed on a 2% agarose gel stained with RedGelTM (Biotium), with 100 volts for 45 min, and DNA bands were visualized with a transilluminator (GelDoc, Biorad). A single band of PCR product indicate the presence of a Mycoplasma spp., and the presence of two bands indicate Acholeplasma spp. (table 2).
TABLE 2 Primers used for the PCR targeting the 16S-23S rARN intergenic spacer regions (IGSR) of Mycoplasma spp. and Acholeplasma spp.
Source: adapted from Boonyayatra et al. (2012b).
The quality control of the PCR was achieved with genomic DNA extracted from Mycoplasma bovis (ATCC 25025) by the tree methods mentioned before, DNA from Streptococcus agalactiae (ATCC 27956) or a free DNA template were used as negative controls in the reaction. The preparation of the DNA extraction and the PCR protocols were performed in two separate rooms of the laboratory to avoid cross-contamination.
Statistical analysis
The bulk tank milk SCC (BTSCC) was transformed in the natural logarithmic scale (LnBTSCC) in order to achieve a normal distribution. The LnBTSCC of dairy farms included in the study for microbiological and molecular analyses (n = 160) and molecular analyses only (n = 60) were compared by a one-way ANOVA with the LnBTSCC values of those farms that were not included (n = 83). The later was done in order to avoid selection bias of the farms in terms of milk quality and subclinical mastitis. Also, a one-way ANOVA was used to determine if there were differences between the farms' LnBTSCC stratified by the region (province) in which they were located or the dairy company they were affiliated to. A P-value of < 0.05 was considered as a statistically significant difference.
Likewise, 95% confidence intervals (95% CI) around the proportion (prevalence) and mean values were calculated (Brown et al. 2001; Corbeil et al. 2019). The statistical analyses were performed in Stata 14 (College Station, Texas USA).
RESULTS
No differences were observed between the LnBTSCC according to the origin of the sample, province, or dairy company. The overall mean LnBTSCC was 6.17 x103 cells/ mL (95% CI = 6.10 - 6.25 x103 cells/mL) for the farms included (microbiology/ molecular analyses and only molecular analyses) in the study and 6.38 x103 cells/ mL (95% CI = 6.26 - 6.51 x103 cells/mL) for excluded farms. The LnBTSCC did not significantly differ among the regions (P > 0.05) (table 3).
TABLE 3 Mean value of the natural logarithmic bulk tank somatic cell count (LnBTSCC x103 cells/mL) of included and excluded farms stratified by the region they were located
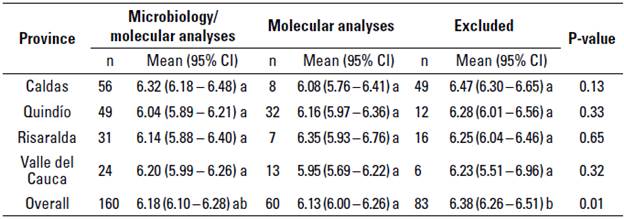
CI, Confidence interval
a-b different superscript letters differ at P < 0.05 between columns in one-way ANOVA and Tukey's post hoc test.
Source: self-made.
During the study none of the 160 farms tested positive to Mycoplasma spp. using microbiological analyses. The presence of suspicious colonies was noted in 3.19% (n = 15) of the samples obtained from 13 farms, but none of them was further identified as Mycoplasma spp. by PCR analysis. The molecular analysis with the genomic DNA extracted directly from the milk shows the same pattern as the obtained via microbiology, where no positive samples were observed. Additional to the latter, the 60 samples of the farms that were included for only PCR, also tested negative to the presence of mycoplasmal DNA in the bulk tank milk. Given all that, and according to the sample size, it is anticipated that the 95% CI for the Mycoplasma spp. prevalence is between 0 and 2.3%, which also indicates with 95% confidence that the prevalence of this pathogen at herd level is < 2.3%.
Fungal or bacteria (mainly gram-negative bacteria) contamination was observed on the 10th day examination in 55.4% (n = 260/459 cultures) of the mycoplasma agar plates. Also, 29.4% (n = 47/160 farms) of the farms showed some contamination on all the cultures of the three samplings at the 10th day of inspection.
DISCUSSION
This was the first study in Colombia that aimed at establishing the prevalence of Mycoplasma spp. at the herd-level based on bulk tank milk samples specifically collected in the provinces of Caldas, Quindío, Risa-ralda, and Valle del Cauca. The later was done according to the Olde Riekerink et al. (2010) recommendations, who concluded that a herd-level prevalence for Mycoplasma spp. should be carried out regionally due to the freshness requirement of bulk tank milk samples and the weak viability of the pathogen for microbiological analysis.
The absence of this pathogen in prevalence studies is not occurring for the first time. Previously, in New Zealand and southeast France there were scientific reports indicating a prevalence for mycoplasma < 1% (McDonald et al. 2009; Arcangioli et al. 2011). However, in New Zealand an outbreak of mycoplasma forced in 2018 the culling of more than 100.000 cows to eradicate the pathogen. In that country, in 2019 (April) the authorities had implemented a mycoplasma eradication program consisting in the analysis of one milk sample once a month during an indefinite period oftime (Dairy NZ 2019).
Moreover, in Israel a 0-3 cases of Mycoplasma bovis mastitis were confirmed per year-herd until 2008. Still, from 2008 on, an increase in the herd level prevalence was observed. Since then 7-9 new cases of Mycoplasma bovis per year-herd were reported, although the cause of this outbreak remains unclear, it could be related to the introduction of a new strain into endemic areas via artificial insemination (Yair et al. 2020). The findings of the latter study reinforce the statement that mycoplasma-associated mastitis is an emerging global problem in the last decade (Fox, 2012).
Despite the results obtained for the mid-western region evaluated, there is enough evidence suggesting that Mycoplasma spp. in absent or low prevalence regions is increasing, and periodical surveillance programs should be established.
Three consecutive samples were analyzed to avoid misdiagnosis and improve the sensitivity of the microbiological analysis for isolating Mycoplasma spp., which is reported for single bulk milk samples between 33 to 59% (Biddle et al. 2003; Olde Riekerink et al. 2006), with a detection limit of of ≥ 1 x101 - 1 x103 CFU/mL (McDonald et al. 2009).
False negative results in the culture may occur due to the lack of viability of the microorganism, the viability of Mycoplasma spp. can be affected by deficient conditions in the manipulation and storage of the samples. In this regard, long time storages, temperature of storage (freezing or/and refrigeration), whether or not additives to cryopreserve samples are used and their concentration (glycerol), inappropriate milk sample handling, and thawing method and temperature, amongst many others (Biddle et al. 2004; Boonyayatra et al. 2010). Due to the above, samples collected in the present research were cultured fresh or within less than 24h after collection and were kept always at a maximum of 4 °C in the meantime. Only samples for molecular analysis were frozen with 30% concentration of cryopreservative added to them.
Additionally, no pre-enrichment of the milk samples was done. Pre-enrichment could have increased the sensitivity of both the microbiological and molecular analyses. Still, this step was not considered because pre-enrichment could also favor the growth of contaminants such as other penicillin-resistant bacterial species (i.e. Acholeplasma landalwii) or fungus present in the sample. This is especially a risk in case of bulk tank milk samples (González and Wilson 2003). However, growth of contaminants is not described as a problem for Mycoplasma spp. growth, recovery and subsequent diagnosis (McDonald et al. 2009). It is also described that pre-enrichment could increase the probability of isolating Mycoplasma bovis by 6%, however, it might increase the cost and time for diagnosis (González and Wilson 2003).
Another factor that could affect the sensitivity of the microbiological analysis is the intermittent shedding pattern of Mycoplasma spp. at individual cow level, that can continue until 13 months after the initial infection (Jasper et al. 1966). For instance, a single cow can shed regularly 1 x103 - 1 x106 CFU/mL of Mycoplasma bovis. Still, those concentrations can also drop below 100 CFU/mL, which is the detection limit described for microbiological identification of Mycoplasma spp. (Biddle et al. 2003; Barkema et al. 2009). Consequently, in case that only one infected cow sheds between 1 x103 to 1 x106 CFU/ mL, a final concentration of Mycoplasma spp. in the bulk milk could be expected between 40 and 4 x104 CFU/mL, assuming that the cows average milk yield is 26 L/day and for herds between 25 and 30 milking cows (Pfützner and Sachse 1996; Arcangioli et al. 2011). In Colombia, the average daily milk yield was 6.3 L/day/cow nationwide (Díaz et al. 2020) and 14 L/day/ cow in the studied regions (Cobo-Ángel et al. 2018), implying that infected cows should shed the Mycoplasma spp. at a much higher concentration in order to be able to detect the pathogen in the bulk milk using microbiological analysis.
Molecular methods were also used for the diagnosis of Mycoplasma spp., because PCR tests are reported to have a high sensitivity and specificity, and can be useful to detect Mycoplasma spp. in milk that was kept frozen up to two years after the sample was collected, thus, a fast and practical alternative to culture (Pinnow et al. 2001; Francoz et al. 2012). According to Arcangioli et al. (2011), PCR could detect Mycoplasma spp. in milk when there is only one cow shedding the pathogen in a herd of 300 milking cows. With a reported detection limit ≤ 5 CFU/mL (Pinnow et al. 2001). In the present study, none of the 210 farms tested positive to the pathogen, even using molecular analysis. This finding supports the results obtained via microbiological analysis of the bulk milk samples.
In the studied regions, there are several studies that revealed a high average of SCC, above 400 x103 cells/mL (Cobo et al. 2015; Cobo-Ángel et al. 2018), which is not different from the values reported in our study. However, those reports also indicate a high prevalence of Staphyloccocus aureus, Streptococcus agalactiae, and Streptococcus uberis as cause for the high BTSCC values. The high BTSCC values obtained in our study can still be attributed to the presence of the abovementioned mastitis causing pathogens on the studied farms.
Assuming that all the due precautions and cares were taken during the study, the lack of Mycoplasma spp. evidence could be explained by several factors related to the production system employed in the country, specifically in the sampled regions. One of them could be the herd size, which can affect the prevalence of mycoplasma caused-mastitis, that rises with increasing the herd size (Fox, 2012). In the sampled regions the herd size average was below 50 lactating cows per herd (Ramírez et al. 2014; Cobo-Ángel et al. 2018), and 85% of the herds had less than 50 cows (ICA, 2019), which can limit the probability of being positive to Mycoplasma spp. Interestingly, in another study conducted in Colombia in a region with a similar average herd size, Mycoplasma spp. were isolated (high-altitude plateau of Boyacá). Still, the Mycoplasma spp. in that study were isolated from quarter milk samples of cows experiencing chronic mastitis that did not respond to antimicrobial treatments and thus not of bulk milk samples (Andrade-Becerra et al. 2014). Furthermore, all evaluated farms in the current study were applying grazing systems, while the main housing types in other studies conducted in the United States, Canada, Chile, and Mexico were free-stall or tie-stall barns (Lemus-Ramírez et al. 2008; Pinedo et al. 2009; Francoz et ail. 2012; Saini et ail. 2013).
Finally, there are some potential protective factors for contagious mastitis including Mycoplasma mastitis in Colombian herds compared to the herds of other countries. One of the most important factors is that the majority of the herds in the region are closed, according to a survey conducted in 2015 (Cobo-Ángel et al. 2018). Not purchasing animals is one of the most effective measures in order to control contagious mastitis pathogens (Keefe 2012). On the other hand, in Colombia most of the farms do not cull cows due to mastitis, which could result in an increased risk and prevalence of contagious mastitis pathogens. However, as it was mentioned before, contagious mastitis pathogens such as Staphylococcus aureus and Streptococcus agalactiae are well-known in the region, while mycoplasma seems to be absent nowadays. Besides, periodic screening programs should be considered at herd level across the national territory to keep Mycoplasma spp. under surveillance, especially in regions where it was previously isolated and from where it can be transferred to other regions through sale or movement of cattle.















