Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Biomédica
Print version ISSN 0120-4157On-line version ISSN 2590-7379
Biomédica vol.27 no.1 Bogotá Jan./Mar. 2007
Parental origin, nondisjunction, and recombination of the extra chromosome 21 in Down syndrome: a study in a sample of the Colombian population
Nelson Javier Ramírez *,1, Helen Marcela Belalcázar *,1, Juan José Yunis 1, 2, Luis Napoleón Quintero 1, 3, Gonzalo Humberto Arboleda 1, 2, Humberto Arboleda 1, 3
1 Grupo de Neurociencias, Instituto de Genética, Universidad Nacional de Colombia, Bogotá D.C., Colombia2 Departamento de Patología, Facultad de Medicina, Universidad Nacional de Colombia, Bogotá D.C., Colombia
3 Departamento de Pediatría, Facultad de Medicina, Universidad Nacional de Colombia, Bogotá D.C., Colombia
*These authors contributed equally to this work
Recibido: 12/09/06; aceptado: 06/02/07
Introduction. Free trisomy 21 is responsible for 95% of Down syndrome cases. Advanced maternal age and susceptible recombination patterns are recognized risk factors associated to Down syndrome. Maternal origin of trisomy occurs in approximately 90% of cases; paternal and mitotic origin share the remaining 10%. However, the recombination events that serve as a risk factors for trisomy 21 have not been carefully characterized.
Objective. To analyze and validate observations in a sample of Colombian trysonomy 21 cases.
Materials and methods. Twenty-two Colombian families were selected, each with one affected Down syndrome (free trisomy 21) child. Microsatellite polymorphisms were used as DNA markers to determine the parental/stage origin of non-disjunction and recombination events. Non-parametric tests were used to compare our results with those reported. Multiple correspondence analysis was used to outline different groups and their associations.
Results. Distribution of trisomy 21 was 90.9% maternal, 4.5% paternal and 4.5% from mitotic origin, similar to distributions reported previously. However, we found differences in the frequency of maternal meiotic stage errors between the present study (46.1% meiosis I and 53.9% meiosis II) compared to those reported previously (70% meiosis I and 30% meiosis II). Multiple correspondence analyses showed association of either local recombination events or absence of recombination with specific non-disjunction stages.
Conclusions. Recombination patterns found in this study support the hypothesis that susceptible chiasmate configurations are associated to maternal meiosis I and meiosis II errors. Non-disjunction frequencies between maternal meiotic stages need to be clarified in our population.
Key words: Down syndrome; nondisjunction, genetic; trisomy; meiosis; recombination, genetic; microsatellite repeats.
Origen parental, estado de no disyunción y recombinación meiótica del cromosoma 21 extra en el síndrome de Down: estudio en una muestra de población colombiana
Introducción. La trisomía 21 libre es responsable del 95% de los casos de síndrome de Down. La edad materna y la recombinación son los principales factores de riesgo asociados con la concepción de estos individuos. El origen materno de la trisomía ocurre en el 90% de los casos, mientras que los casos de origen paterno y mitótico comparten un 10%. Por otra parte, la recombinación como factor de riesgo para la trisomía 21 no ha sido comprobada completamente.
Objetivo. Analizar y validar estas observaciones en una muestra colombiana de casos con trisomía 21 libre.
Materiales y métodos. Se estudiaron 22 afectados con síndrome de Down (trisomía libre) y sus respectivos padres. Se usaron marcadores microsatélites de ADN para determinar el origen en los progenitores, el estado de no disyunción y los eventos de recombinación. Por medio de pruebas no paramétricas se compararon los resultados con los reportados en la literatura. Se realizó análisis de correspondencias múltiples para reconocer los diferentes grupos y sus asociaciones.
Resultados. La distribución de la trisomía 21 fue 90,9% materna, 4,5% paterna y 4,5% mitótica, similar a las reportadas previamente. Sin embargo, existen diferencias en la frecuencia de errores del estado meiótico para origen materno (46,1% meiosis I y 53,9% meiosis II) comparada con la de los reportados previamente (70% meiosis I y 30% meiosis II). El análisis de correspondencias múltiples mostró asociación entre eventos de recombinación local y ausencia de recombinación con estados de no disyunción específicos.
Conclusiones. Las configuraciones quiasmáticas susceptibles están asociadas de manera específica a errores en la meiosis I y la meiosis II materna. Es necesario clasificar la frecuencia de no disyunción en estados meióticos maternos en nuestra población.
Palabras clave: síndrome de Down, no disyunción genética, trisomía, meiosis, recombinación genética, repeticiones de microsatélite.
Trisomy 21 (Down syndrome) is the most commonly occurring supernumerary chromosome anomaly leading to mental retardation. The extra chromosome 21 arises as a consequence of non-disjunction during meiosis in gametes or during postzygotic mitosis.
Down syndrome prevalence rates differ by ethnicity and culture. Hispanics have higher rates of Down syndrome than other genetic backgrounds (1,2). For instance, an incidence of 1 in 700 has been reported for Hispanics living in the USA, less than 1 in 600 in several non-related Latin-American populations, 1 in 1,075 for USA Caucasians, and 1 in 700 to 1 in 1,000 for other Caucasian populations (3-6). These rates are also affected by the number of pregnancies terminated as a consequence of prenatal screening.
Recent evidence indicated a significantly greater risk of Down syndrome in live and stillbirths among Hispanic women than those of Caucasian women (7). Additionally, a higher risk of Down syndrome was found in foreign-born Hispanic women [1,7] compared to Hispanic women and Caucasian non-Hispanic women born in the United States [1,2]. Carothers et al. suggested an increased rate of Down syndrome among US residents of Mexican or Central American origin (8), but these rates were not corroborated in a study that included data from nine South American countries in a non-population based study (9).
Colombia, which is composed mainly of Caucasian-Mestizo and African descendent populations (10), has a Down syndrome incidence around 1 in 600 (11,12) and recently was estimated as 1 in 578 (Ignacio Manuel Zarante, ECLAM, personal communication).
Several studies have used DNA polymorphisms to identify the origin of the extra chromosome 21 (13-15). In the largest meta-analysis study of 807 Down syndrome patients, the parental origin was maternal in 90.7% of cases, paternal in 5.5% and mitotic in the remaining 3.8% (16). Additionally, when pericentromeric microsatellite markers were used to detect the meiotic stage of non-disjunction, 76% of maternal cases were found to occur at meiosis I (MI) and 24% at meiosis II (MII); in the cases of paternal origin, a 1:1 ratio between stages was found, with a slight excess of MII errors (13).
Recombination is a second important source of non-disjunction (after maternal age) and is the only subcellular event related to this process in humans (17,18). The associations between (a) specific meiotic errors and (b) absence of or localized recombination suggested that all non-disjunction events are initiated during MI and then resolved at either MI or MII (19).
In the current study, the parental origin, the meiotic/mitotic stage of origin, and the effect of recombination events on the production of trisomy 21 are determined in a sample of Colombian families with Down syndrome children.
Materials and methods
Sampling
The population sample consisted of 22 young Colombian probands clinically diagnosed and cytogenetically verified as Down syndrome patients (free trisomy 21), as well as their progenitors. All families were living in Bogotá, but had immigrated from different parts of Colombia and were analyzed at Instituto de Genética, Universidad Nacional de Colombia. The average age of Down syndrome children was 3.2 years (range 0–18). Average maternal age at the time admission to the study was 31.5±10.2 years, and average paternal age was 35.7±14.0 years. Parental age at the time of the patients medical examination was considered in order to assess the temporal environmental conditions for possible risk factors during Down syndrome conceptions. Informed consent was obtained from parents in all cases. All the procedures were in accordance with the ethical standards of the Universidad Nacional de Colombia Ethical Committee on Human Experimentation and with the Helsinki Declaration (20).
DNA studies
High molecular-weight genomic DNA was extracted from peripheral blood cells from probands and their parents. Six microsatellite polymorphisms of chromosome 21 were isolated using PCR amplification and separated by PAGE (polyacrylamide gel electrophoresis). The set of microsatellites, arrayed between the centromere and telomere of the long arm of chromosome 21, included: D21S1432, D21S11, D21S1437, APP Intron 7, D21S1411, and PFKL (
Figure 1).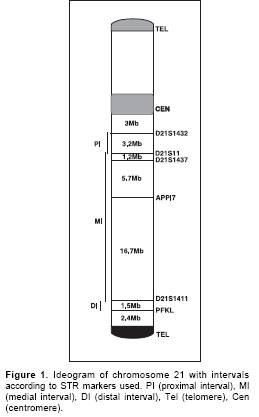
PCR amplification was carried out in 25µl final volume as described elsewere (21); annealing temperatures for each microsatellite marker were according with manufacturer recommendations. PAGE was carried out in 6-8% acrylamide–bis-acrylamide denaturing gels depending on fragment sizes for each STR (short tandem repeat). Gels were silver nitrate stained (
Figure 2).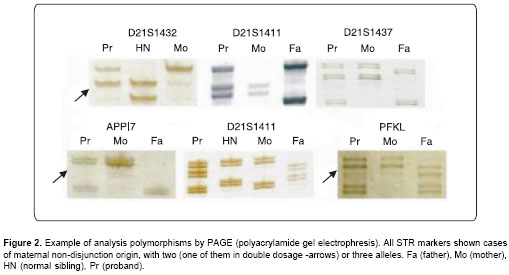
Definition of the parental origin, the meiotic/mitotic stage of origin and recombination events
Parental origin was determined by the results from at least two STR markers using digital dosage analysis with Scion Image (version Beta 4.0.2). The determination was possible when two variant alleles were present in the proband, or by comparing the polymorphic alleles when three different alleles were present in the proband. Subsequently, the meiotic stage of origin of trisomy was determined by the state of reduction in chromosome 21 pericentromeric markers of the parent who had contributed the extra chromosome. If parental heterozygosity was retained in the trisomic offspring, the error occurred during MI, and if parental heterozygosity was reduced to homozygosity, the error occurred during MII or mitosis. Mitotic errors were distinguished from MII errors by evaluating non-pericentromeric markers. If a trisomic individual was reduced to homozygosity at the informative loci, a post-zygotic origin was inferred. If the trisomic individual was not reduced to homozygosity at one or more loci, a MII origin was indicated.
For recombination analysis, the 21q chromosome was treated in three recombinational intervals as follows: D21S1432-D21S11 was the proximal interval, D21S11-D21S1411 was the medial interval, and D21S1411-PFKL was the distal interval (Figure 1).
Statistical analysis
A chi-squared test for k proportions was used to compare parental origin and meiotic stage with those reported elsewhere (22). Students t-test was used to compare mean ages between maternal subgroups.
We used multiple correspondence analyses (MCA) to qualitatively detect factors influencing maternal non-disjunction. The MCA represented categories as points in a multidimensional space. The distance between points, defined according to a chi-square metric, expressed the dissimilarities between categories. MCA provided a low-dimensional picture that approximated the structure of the cluster of points in the space identified in the first factorial axes, which were the axes of maximum dispersion of the cluster. For graphical illustration, only the two axes providing the largest amount of information were used. MCA was performed only for 13 cases of maternal origin with known meiotic stage. Meiotic stage, recombination position, maternal and paternal age at child birth, proband sex, and parity at Down syndrome child birth were the categorical variables. A contingency table was constructed, including the results for each variable and for each individual. The MCA was made with the Statistical Package for Augmented Designs (SPAD) (version 5.5).
Results
Parental origin and meiotic stage were determined for 22 probands with Down syndrome, consistint of 15 males and 7 females. Mean maternal age at proband birth was 28.1 years (range 14-41 years) and mean paternal age was 34.1 years (range 18-52 years).
Parental origin was determined in all cases. Maternal origin was determined in 20 cases (90.9%), paternal in one case (4.6%) and mitotic in one case (4.6%) (Table 1). Meiotic or mitotic origin of the extra chromosome 21 was identified in 14 of 22 cases. Six cases were consistent with MI error, seven with MII error, and one with mitotic error (Table 1). All meiotic cases (MI: 46,15%; MII: 53,87%) were maternal in origin. The mean and the standard deviation of maternal age (at proband birth) in families with trisomy of maternal origin, but unknown meiotic origin, was 27.8±8.0 years; in families with maternal MI errors the age was 22.8±7.7 years; and in families with maternal MII errors the age was 27.7±8.0 years. Not statistically significant differences were seen among the maternal ages.
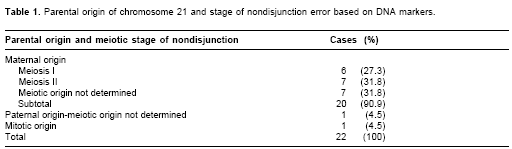
The number and location of crossovers were obtained for disjoined chromosomes of maternal MI cases (n=6) and maternal MII cases (n=7) (Table 2). Distal positioning exchange (one case) and possible achiasmate chromosomes (five cases) were noted for maternal MI cases. In maternal MII cases, recombination events occurred in the proximal (four cases) and medial intervals (three cases). Additionally, single recombination events were found in three undetermined maternal cases.
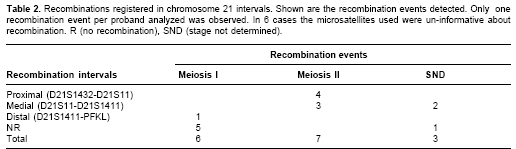
Graphical representation of MCA (Figure 3) shows that the better scattered variables along both dimensions (factor axes) were recombination, meiotic stage of non-disjunction and maternal age. This analysis separated two groups of factors: (1) MII, medial or proximal recombination, and mothers at least 29 years old (at proband birth) were associated, and (2) MI, distal or possibly no recombination and mothers with less than 29 years. Parity (Down syndrome position), proband gender and paternal age did not seem associated with any of the groups.
Discussion
The results of parental origin of trisomy 21 were not different from those already reported (16,18,23). Warren et al. first suggested that reduced chiasma formation along chromosome 21 predisposes it to non-disjunction errors (24). The above data suggest two clusters (Figure 3, dashed ovals), the first is the MI-No recombination-distal recombination cluster and the second is the MII-Proximal recombination-medial recombination cluster. Presence of these clusters support the hypothesis that almost all non-disjunction cases are the result of errors initiated at MI (19,25).
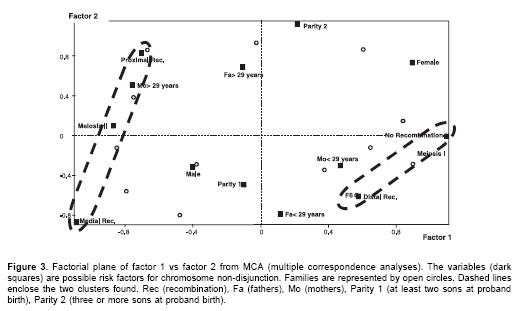
In maternal cases, the stage of non-disjunction was determined in 13 of 20 cases. Seven were at MI, and 8 at MII. These data differ significantly (p<0.01) from those previously described (16). These differences may have diverse explanations: first, a reduced sample size have led to random statistical variation rather than to a real effect; second, an increment in true MII errors (those who are not associated with proximal recombination) raises the number of MII cases. Third, an increase in non-disjunction events associated with proximal recombination events recorded as "MII", as seen the cause in the present results. Despite of the small sample size, the proportions of parental origin data and recombination patterns were similar to those found in other studies and they seem not be affected by random. On the other hand, studies with "reduced" sample size did not found differences analyzing meiotic stage in trisomies compared with those reported for large samples (14,26,27)
Since cases of proximal recombination and medial recombination are equally associated to MII errors (Figure 3), the other alternatives cannot be eliminated. One explanation is that non-disjunction has multifactorial influences, where age plays a role in some cases but not in others, and where other influencing factors act globally in cells or locally in chromosomes (17). Recombination is also a multifactorial trait affecting chromosome segregation, and it has been reported that alteration of crossover patterns in gametes is associated with non-disjunction (17,28). To determine whether recombination-associated factors or independent factors are responsible for these results, further investigation is required.
In the current study, mothers less than 29 years old (at the childs birth) clustered with MI errors, and mothers older than 29 years clustered with MII errors. Although no significant differences in maternal mean age were detected between MI and MII cases, this tendency is a constant in others studies (16,18,23).
The molecular correlates of non-disjunction in oocytes involve several mechanisms: chromosome condensation (29,30), pairing of homologous chromosomes (31,32), synaptonemal complex formation (33,34), recombination events (35,36), spindle formation (37,38) and meiotic checkpoints (39,40). Proteins actively functioning in these processes are essential for proper development, but their specific roles in oocyte maturation are difficult to unravel. The "two hit" model proposed in 1996 (25) and extended recently (41) offer an alternative explanation to non-disjunction cases involving recombination. Susceptible recombinants generate a first hit, and then a second hit, related to mother age, triggers non-disjunction events. Cases in which recombination did not generate susceptible configurations possibly involve subtle modifications (for instance, SNP coding variations for effector proteins) in any of the mechanisms listed above.
In summary, the Colombian sample indicates an interesting population which free trisomy 21 allocates non-disjunction events in different proportion to the meiotic stages. To substantiate these observations, additional studies in other populations with a similar background must be undertaken. Additional epidemiological and molecular studies will be important in order to interpret these results within the framework of a possible high incidence of Down syndrome in Colombia.
Acknowledgements
We wish to thank Dalila Camelo for cytogenetic study of some samples and Diana Rincon for her advice in statistic methods.
Conflict of interest
None declared.
Grant support
This work was supported by grants from DIB-Universidad Nacional de Colombia (822018, 209002 and 807080) and Colciencias (1101-04-10161-2000).
Corresponding: Humberto Arboleda, Profesor Asociado, Departamento de Pediatría, Facultad de Medicina, Universidad Nacional de Colombia, Carrera 30 No.45-03, Bogotá D.C., Colombia.
Tel:3165000 ext. 11613, fax: 3165526, harboledag@unal.edu.co
References
1. Chavez GF, Cordero JF, Becerra JE. Leading major congenital malformations among minority groups in the United States, 1981-1986. MMWR CDC Surveill Summ 1988;37:17-24. [ Links ]
2. Center for Disease Control and Prevention. Down syndrome prevalence at birth-United States, 1983-1990. MMWR Morb Mortal Wkly Rep 1994;43:617-22. [ Links ]
3. Bishop J, Huether CA, Torfs C, Lorey F, Deddens J. Epidemiologic study of Down syndrome in a racially diverse California population, 1989-1991. Am J Epidemiol 1997;145:134-47. [ Links ]
4. Nazer J, Margozzini J, Rodriguez M, Rojas M, Cifuentes L. Disabling malformations in Chile. Latin American Cooperative Study of Congenital Malformations (ECLAMC), 1982-1997. Rev Med Chil 2001;129:67-74. [ Links ]
5. Roizen NJ, Patterson D. Downs syndrome. Lancet 2003;361:1281-9. [ Links ]
6. Ojeda ME, Moreno R. High prevalence of Down syndrome in the Rancagua Hospital in central Chile. Rev Med Chil 2005;133:935-42. [ Links ]
7. Carmichael SL, Shaw GM, Kaidarova Z. Congenital malformations in offspring of Hispanic and African-American women in California, 1989-1997. Birth Defects Res A Clin Mol Teratol 2004;70:382-8. [ Links ]
8. Carothers AD, Hecht CA, Hook EB. International variation in reported livebirth prevalence rates of Down syndrome, adjusted for maternal age. J Med Genet 1999;36:386-93. [ Links ]
9. Carothers AD, Castilla EE, Dutra MG, Hook EB. Search for ethnic, geographic, and other factors in the epidemiology of Down syndrome in South America: analysis of data from the ECLAMC project, 1967-1997. Am J Med Genet 2001;103:149-56. [ Links ]
10. Yunis JJ, Acevedo LE, Campo DS, Yunis EJ. Population data of Y-STR minimal haplotypes in a sample of Caucasian-Mestizo and African descent individuals of Colombia. Forensic Sci Int 2005;151:307-13. [ Links ]
11. Ramírez RE, Isaza C, Gutiérrez MI. La incidencia del síndrome de Down en Cali. Colomb Med 1996;27:138-42. [ Links ]
12. Castilla EE, Lopez-Camelo JS, Campana H. Altitude as a risk factor for congenital anomalies. Am J Med Genet 1999;86:9-14. [ Links ]
13. Savage AR, Petersen MB, Pettay D, Taft L, Allran K, Freeman SB, et al. Elucidating the mechanisms of paternal non-disjunction of chromosome 21 in humans. Hum Mol Genet 1998;7:1221-7. [ Links ]
14. Gomez D, Solsona E, Guitart M, Baena N, Gabau E, Egozcue J et al. Origin of trisomy 21 in Down syndrome cases from a Spanish population registry. Ann Genet 2000;43:23-8. [ Links ]
15. Diego-Alvarez D, Garcia-Hoyos M, Trujillo MJ, Gonzalez-Gonzalez C, Rodriguez de AM, Ayuso C et al. Application of quantitative fluorescent PCR with short tandem repeat markers to the study of aneuploidies in spontaneous miscarriages. Hum Reprod 2005;20:1235-43. [ Links ]
16. Petersen MB, Mikkelsen M. Nondisjunction in trisomy 21: origin and mechanisms. Cytogenet Cell Genet 2000; 91:199-203. [ Links ]
17. Lamb NE, Sherman SL, Hassold TJ. Effect of meiotic recombination on the production of aneuploid gametes in humans. Cytogenet Genome Res 2005;111:250-5. [ Links ]
18. Sherman SL, Freeman SB, Allen EG, Lamb NE. Risk factors for nondisjunction of trisomy 21. Cytogenet Genome Res 2005;111:273-80. [ Links ]
19. Lamb NE, Feingold E, Savage A, Avramopoulos D, Freeman S, Gu Y et al. Characterization of susceptible chiasma configurations that increase the risk for maternal nondisjunction of chromosome 21. Hum Mol Genet 1997;6:1391-9. [ Links ]
20. World Medical Association. World Medical Association Declaration of Helsinki. Ethical principles for medical research involving human subjects. JAMA 2000;284:3043-5. [ Links ]
21. Forero DA, Benitez B, Arboleda G, Yunis JJ, Pardo R, Arboleda H. Analysis of functional polymorphisms in three synaptic plasticity-related genes (BDNF, COMT AND UCHL1) in Alzheimers disease in Colombia. Neurosci Res 2006;55:334-41. [ Links ]
22. Gibbons J, Chakraborty S. Nonparametric Statistical Inference. 3rd. ed. New York: Marcel Dekker; 1992. [ Links ]
23. Ballesta F, Queralt R, Gomez D, Solsona E, Guitart M, Ezquerra M et al. Parental origin and meiotic stage of non-disjunction in 139 cases of trisomy 21. Ann Genet 1999;42:11-5. [ Links ]
24. Warren AC, Chakravarti A, Wong C, Slaugenhaupt SA, Halloran SL, Watkins PC et al. Evidence for reduced recombination on the nondisjoined chromosomes 21 in Down syndrome. Science 1987; 237:652-4. [ Links ]
25. Lamb NE, Freeman SB, Savage-Austin A, Pettay D, Taft L, Hersey J et al. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet 1996;14:400-5. [ Links ]
26. Chen CP, Chern SR, Tsai FJ, Lin CY, Lin YH, Wang W. A comparison of maternal age, sex ratio and associated major anomalies among fetal trisomy 18 cases with different cell division of error. Prenat Diagn 2005;25:327-30. [ Links ]
27. Vorsanova SG, Iourov IY, Beresheva AK, Demidova IA, Monakhov VV, Kravets VS et al. Non-disjunction of chromosome 21, alphoid DNA variation, and sociogenetic features of Down syndrome. Tsitol Genet 2005;39:30-6. [ Links ]
28. Brown AS, Feingold E, Broman KW, Sherman SL. Genome-wide variation in recombination in female meiosis: a risk factor for non-disjunction of chromosome 21. Hum Mol Genet 2000;9:515-23. [ Links ]
29. Dej KJ, Ahn C, Orr-Weaver TL. Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics 2004; 168:895-906. [ Links ]
30. Resnick TD, Satinover DL, Macisaac F, Stukenberg PT, Earnshaw WC, Orr-Weaver TL et al. INCENP and Aurora B Promote Meiotic Sister Chromatid Cohesion through Localization of the Shugoshin MEI-S332 in Drosophila. Dev Cell 2006;11:57-68. [ Links ]
31. Martinez-Perez E, Villeneuve AM. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev 2005;19:2727-43. [ Links ]
32. Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 2006;125:59-69. [ Links ]
33. Yuan L, Liu JG, Hoja MR, Wilbertz J, Nordqvist K, Hoog C. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 2002;296:1115-8. [ Links ]
34. Wang H, Hoog C. Structural damage to meiotic chromosomes impairs DNA recombination and checkpoint control in mammalian oocytes. J Cell Biol 2006;173:485-95. [ Links ]
35. Tease C, Hartshorne GM, Hulten MA. Patterns of meiotic recombination in human fetal oocytes. Am J Hum Genet 2002;70:1469-79. [ Links ]
36. Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, Poulos BK et al. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis I in human oocytes. Am J Hum Genet 2005;76:112-27. [ Links ]
37. Cullen CF, Brittle AL, Ito T, Ohkura H. The conserved kinase NHK-1 is essential for mitotic progression and unifying acentrosomal meiotic spindles in Drosophila melanogaster. J Cell Biol 2005;171:593-602. [ Links ]
38. Pearson NJ, Cullen CF, Dzhindzhev NS, Ohkura H. A pre-anaphase role for a Cks/Suc1 in acentrosomal spindle formation of Drosophila female meiosis. EMBO Rep 2006;6:1058-63. [ Links ]
39. Hochwagen A, Tham WH, Brar GA, Amon A. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintainmeiotic recombination checkpoint activity. Cell 2005;122:861-73. [ Links ]
40. Homer HA, McDougall A, Levasseur M, Yallop K, Murdoch AP, Herbert M. Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securin during meiosis I in mouse oocytes. Genes Dev 2005;19:202-7. [ Links ]
41. Warren WD, Gorringe KL. A molecular model for sporadic human aneuploidy. Trends Genet 2006; 22:218-24. [ Links ]














