Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Biomédica
Print version ISSN 0120-4157On-line version ISSN 2590-7379
Biomédica vol.27 no.2 Bogotá Apr./June 2007
In vitro
mating of Colombian isolates of the Cryptococcus neoformans species complexPatricia Escandón 1, Popchai Ngamskulrungroj 2, Wieland Meyer 2, Elizabeth Castañeda 1
1 Grupo de Microbiología, Instituto Nacional de Salud, Bogotá, D. C., Colombia
2
Molecular Mycology Research Laboratory, Center for Infectious Diseases and Microbiology, The University of Sydney Western Clinical School at Westmead Hospital/Westmead Millennium Institute, Westmead, NSW, AustraliaRecibido: 15/02/07; aceptado: 30/04/07
Introduction.
Within the Cryptococcus neoformans species complex, two species and five serotypes are recognized: C. neoformans (var. grubii, serotype A; var. neoformans, serotype D and a hybrid, serotype AD) and C. gattii (serotypes B and C). Mating types a and a are designated by a single locus, with the mating type a being most prevalent in serotype A and D strains.Objective. To evaluate the ability of Colombian isolates of the C. neoformans species complex to mate in vitro with tester strains of the opposite mating type.
Materials and methods. Fifty three clinical isolates were included in this study, 33 C. neoformans var. grubii serotype A, 4 C. neoformans var. neoformans serotype D, all mating type
a, and 16 C. gattii, 13 serotype B (mating type a) and 3 serotype C (mating type a), were mixed on V8 juice agar, using a modified method, with the appropriate tester strains to determine the mating types in vitro.Results. Mating studies revealed that 9 of 33 (27.3%) serotype A isolates and 6 of 13 (46.2%) serotype B isolates were able to mate. Clamp connections and basidia with basidiospores were observed microscopically, indicating that the mating process had occurred. All mating competent serotype A strains were mating type alpha and the serotype B mating competent strains were mating type a.
Conclusion. This is the first report of the determination of the mating ability of Colombian Cryptococcus neoformans isolates to mate in vitro with appropriate tester strains, which is of great importance to study the propagation of the fungus around the globe.
Key words: Cryptococcus neoformans, phenotype, in vitro, genes, fungal
Determinación in vitro de la pareja sexual en aislamientos del complejo Cryptococcus neoformans
Introducción. En el complejo Cryptococcus neoformans se reconocen dos especies y cinco serotipos: C. neoformans (var. grubii, serotipo A; var. neoformans, serotipo D y un híbrido, serotipo AD) y C. gattii (serotipos B y C). La pareja sexual a y a es controlada por un solo locus, y la pareja sexual a es la más prevalente en los serotipos A y D, y es convencionalmente determinada mediante reacción en cadena de la polimerasa.
Objetivo. Evaluar la habilidad de aislamientos colombianos de C. neoformans para aparearse in vitro con aislamientos control de la pareja sexual opuesta.
Materiales y métodos. Treinta y tres aislamientos clínicos de C. neoformans var. grubii serotipo A, 4 de la var. neoformans serotipo D, todos pareja sexual a, y 16 aislamientos clínicos de C. gattii, 13 serotipo B (pareja sexual a) y 3 serotipo C (pareja sexual a ), se mezclaron, en agar jugo V8 modificado, con cepas control para determinar la pareja sexual in vitro.
Resultados. Los estudios de apareamiento mostraron que 9 de 33 (27,3%) aislamientos serotipo A y 6 de 13 (46,2%) aislamientos serotipo B tuvieron la capacidad de aparearse con las cepas control. Todos los aislamientos del serotipo A que presentaron apareamiento eran pareja sexual a y los del serotipo B eran pareja sexual a. Microscópicamente se observaron conexiones en gancho, basidias y basidiosporas, estructuras que establecieron que se había realizado el proceso de apareamiento.
Conclusión. Este acercamiento provee por primera vez la capacidad de que los aislamientos colombianos de C. neoformans se apareen in vitro con cepas control lo cual tiene importancia en el estudio de la diseminación del hongo.
Palabras clave: Cryptococcus neoformans, fenotipo, in vitro, genes, hongos
Isolates of theCryptococcus neoformans species complex cause cryptococcosis, the second most common life-threatening invasive fungal disease in humans and animals worldwide. Cryptococcosis is a systemic mycoses that affects immunocompetent and immunosuppressed individuals (1).
The C. neoformans species complex contains two closely related species. The first species: C. neoformans, which includes two varieties: C. neoformans var. grubii (serotype A, genotypes VNI/AFLP1 and VNII/AFLP1A) and C. neoformans var. neoformans (serotype D, genotype VNIV/AFLP2) as well as an AD hybrid (genotype VNIII/AFLP3), is an opportunistic pathogen that typically causes disseminated cryptococcosis in hosts with normal or impaired immunity (2-4), C. neoformans var. grubii is the major fungal pathogen in patients with AIDS (1,2). The second species: C. gattii (serotypes B and C, genotypes VGI/AFLP4, VGII/AFLP6, VGIII/AFLP5 and VGIV/AFLP7) is a primary pathogen that mainly affects patients with normal immunity (3-5). Recently, a natural occurring hybrid was described between C. neoformans and C. gattii serologically characterized as BD (6). These serotypes/molecular types differ in their epidemiological, ecological, virulence and molecular characteristics (1, 4,7-11). Infections are assumed to be acquired via inhalation of infectious propagules, assumed to be desiccated yeast cells (blastoconidia) or basidiospores, from environmental niches (1,7,8).
In the C. neoformans species complex, the mating system is controlled by a single locus with two functional alleles, which designate the mating types MATa and MATa (12). The mating type locus has been associated with virulence, as suggested by Kwon-Chung et al. (13), who found that MATa strains were more virulent in a mouse model than the MAT aisolates. Recently it was found that the cryptococcosis outbreak on Vancouver Island, Canada was caused by isolates that were highly virulent mating type astrains belonging to the C. gattii molecular type VGII/AFLP6 (14,15).
Epidemiological surveys have shown that mating type a strains are most prevalent in serotype A isolates, both clinical and environmental (16), whereas in serotype B, we found that mating type a is more frequent in Colombia (17).
In our laboratory, the mating type has been determined by PCR using mating type specific primers within the MF1a/MF1a genes, which encode pheromones (16,18). The ability of C. neoformans strains to mate is determined by crossing the isolates with appropriate tester strains on nutrient starvation media (1,14,16). The tester strains (usually JEC 20, serotype D, mating type a, and JEC21, serotype D, mating type a) are mixed with the strains to be tested on a special media like V8 agar, SYB (sucrose, biotin, yeast extract) (19). Strains with different mating types mate to form dikaryotic hyphae, clamp connections, basidia and basidiospores, these structures are indicative of compatible mating types (20). However, the determination of the mating ability in vitro is associated with a number of problems, like the fact that mating is sensitive to a variety of environmental conditions, including temperature, nutrient availability, and moisture levels (1), and many strains, loose their ability to mate after being extensively manipulated (16). Despite these problems, the determination of the mating potential in vitro is of great interest because sexual recombination is important for the dissemination and propagation of the fungus in the environment and its transition into humans and animals (21).
The aim of the present study was to evaluate the ability of Colombian isolates of the C. neoformans species complex to mate in vitro with tester strains and produce structures associated with the mating process.
Materials and methods
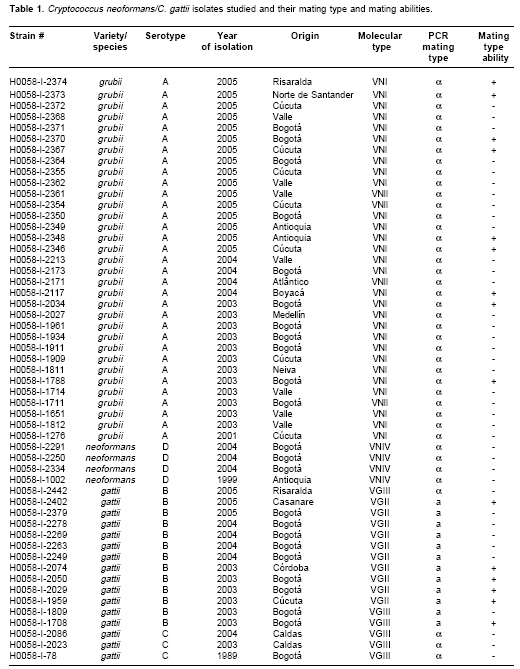
Fungal isolates. Fifty three clinical isolates of C. neoformans and C. gattii recovered between 1989 and 2005 were studied. From these isolates, 33 were C. neoformans var. grubii serotype A, 4 were C. neoformans var. neoformans serotype D, 13 isolates were C. gattii serotype B and 3 were serotype C, as seen in table 1. The serotype was previously determined using the Crypto-Check, Iatron Laboratories, Tokyo, Japan. The mating type was previously determined by PCR, using MATa and MATa specific primers corresponding to the MF1aand MF1a genes (16), 40 isolates were mating type a and 13 were mating type a(table 1). All isolates originated from Colombia. The isolates were maintained as glycerol stocks at -70°C for long-term storage.
Reference strains
. Strains H0058-I-1127 (JEC 20, serotype D, mating type a) and H0058-I-1128 (JEC 21, serotype D, mating type a), kindly provided by June Kwon Chung from the National Institutes of Health in Bethesda, USA, were used as tester strains for the mating (table 1).Determination of in vitro mating. Each strain was crossed with the opposite mating partner tester strain, using a slightly modified method originally described by Kwon Chung et al. (22). Briefly, each strain was grown on Sabouraud agar at 27°C for 24 h. Then a tiny amount of each strain was taken using a toothpick and streak crossed each other, in a very thin layer, on V8 juice agar (5% V8 canned juice (Campbell´s), 3mM KH2PO4, 4% agar) adjusted to pH 5.0 with KOH 1N. Plates were incubated in darkness at 25°C for at least 4 weeks in a dry place, without wrapping the plate. Plates were examined regularly for evidence of hyphae with clamp connections, basidium, basidia and basidiospore chains indicative of mating using a microscope.
Results
Mating was observed in 15 isolates studied, from which 9 were serotype A and 6 serotype B, that mated with the opposite mating partner tester strain. As a result of the mating all mating competent serotype A strains were mating type
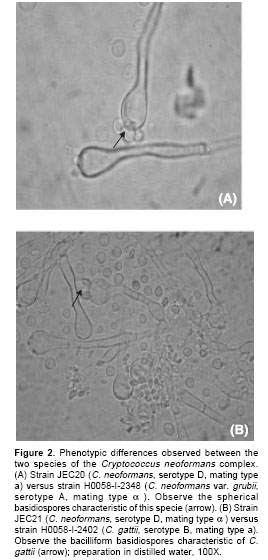
a and the mating competent serotype B strains were mating type a, confirming the results obtained previously using PCR amplification of the MF1a/MF1a genes with MATa and MATaspecific primers.A first sign that the mating process had occurred was the presence of filaments on the agar where the strains were mixed. Afterwards, microscopic evidence showed the presence of fungal structures (mycelium, clamp connections) compatible with mating in vitro. After conjugation had occurred, a dikaryotic mycelium with clamp connections was observed (
figure 1A), followed by the formation of terminal basidia with basidiospores (figure 1B), which then germinate into an encapsulated yeast cell. Phenotypic differences were observed microscopically between the two species: the basidiospores of C. neoformans var. grubii were spherical or cylindrical (figure 2A), whereas those of C. gattii were bacilliform (figure 2B).
Discussion
The present study described the in vitro mating process between isolates of the C. neoformans species complex on V8 agar using a slightly modified protocol. The fact that each competent strain, which mating type had been previously determined by PCR, mated with the opposite tester strain, reveals a 100% concordance with the PCR. This is the first time in our laboratory, that we were able to successfully mate isolates of opposite mating types with reproducible results, since the experiment was done twice, with the same results. Several approaches had been made previously in our laboratory to test those strains, including culture on SYB agar and malt agar (22), but those experiments had been not reproducible using the previously described methodologies and the mating experiments had to be repeated more than once (23).
The sexual stage has not been conclusively identified in nature although there has been some suggestion that structures of C. gattii might be found in flowers from eucalyptus trees (7); however, under conditions of nitrogen starvation and relative desiccation, cells of the two mating types in physical proximity can conjugate and form a dikaryotic mycelium in the laboratory (1) as observed in our crosses that mated successfully when culturing the isolates on V8 media using a modified method.
It is interesting to observe that the mating process occurred in isolates of both mating types in a similar ratio, 9:6 (a:a), taking into account that it had been reported, that the a mating type strains occur in over 90% of the worldwide C. neoformans clinical and environmental isolates (14,24). However, it was recently reported by our group that 96.6% of serotype B isolates are of mating type a (17).
It is also remarkable to show that all of our serotype B isolates, which were able to mate in vitro, are of mating type a, which is interesting taking into consideration that all the isolates that produced the outbreak on Vancouver Island were of mating type a(14). The herein standardized technique will encourage us to mate the Colombian strains with those that belong to the Vancouver Island outbreak, which will help to elucidate how isolates of the C. neoformans species complex could have evolved into more virulent strains, which subsequently may have caused an outbreak outside the natural habitat of C. gattii.
Conflict of interests
The authors of the present article declare that there are no conflicts of interest that may have influenced the results of this work.
Financing
This work was supported by the Instituto Colombiano para el Desarrollo de la Ciencia y la Tecnología "Francisco José de Caldas", Colciencias (código 2104-05-14656) and the Instituto Nacional de Salud.
Correspondencia:
Patricia Escandón, Grupo de Microbiología, Instituto Nacional de Salud, Avenida calle 26 No. 51-20, Bogota, Colombia.
Teléfono: 57-1-2207700 ext. 433; fax: 57-1-2207700 ext.445
pescandon@ins.gov.co
References
1. Casadevall A, Perfect JR. Cryptococcus neoformans. Washington, D.C.: American Society for Microbiology Press; 1998. p.541. [ Links ]
2. Franzot SP, Salkin IF, Casadevall A. Cryptococcus neoformans var. grubii: a separate varietal status for Cryptococcus neoformans serotype A isolates. J Clin Microbiol. 1999;37:838-40. [ Links ]
3. Boekhout T, Theelen B, Diaz M, Fell JW, Hop WC, Abeln EC, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891-907. [ Links ]
4. Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardke C, Methling K, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA- a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20:1790-9. [ Links ]
5.Kwon-Chung K, Boekhout T, Fell J, Diaz M. Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon. 2002; 51:804-6. [ Links ]
6. Bovers M, Hagen F, Kuramae EE, Diaz MR, Spanjaard L, Dromer F, et al. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6: 599-607. [ Links ]
7. Ellis DH, Pfeiffer TJ. Natural habitat of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1990;28:1642-4. [ Links ]
8. Sorrell TC, Ellis DH. Ecology of Cryptococcus neoformans. Rev Iberoam Micol. 1997;14:42-3. [ Links ]
9. Meyer W, Castañeda A, Jackson S, Huynh M, Castañeda E, Iberoamerican Cryptococcal Study Group. Molecular typing of Iberoamerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9:189-95. [ Links ]
10. Stephen C, Lester S, Black W, Fyfe M, Raverty S. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can Vet J. 2002;43:792-4. [ Links ]
11. Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA, Heitman J. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol. 2005;3:753-64. [ Links ]
12. Kwon-Chung KJ. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976;68:821-33. [ Links ]
13. Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602-5. [ Links ]
14. Kidd SE, Hagen F, Tscharke LR, Huynh M, Bartlett KH, Fyfe M, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci USA. 2004;101:17258-63. [ Links ]
15. Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S et al. Same sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360-4. [ Links ]
16. Halliday CL, Bui T, Krockenberger M, Malik R, Ellis DH, Carter DA. Presence of a and a mating types in environmental and clinical collections of Cryptococcus neoformans var. gattii strains from Australia. J Clin Microbiol. 1999;37:2920-6. [ Links ]
17. Escandón P, Sánchez A, Martínez M, Meyer W, Castañeda E. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res. 2006;6:625-35. [ Links ]
18. Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell 2003;2:1036-45. [ Links ]
19. Kwon-Chung KJ. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:942-6. [ Links ]
20. Wickes BL, Mayorga ME, Edman U, Edman JC. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the a mating type. Proc Natl Acad Sci USA. 1996;93:7327-31. [ Links ]
21. Halliday CL, Carter DA. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J Clin Microbiol. 2003;41:703-11. [ Links ]
22. Kwon-Chung KJ, Bennett JE, Rhodes JC. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Van Leeuwenhoek. 1982;48:25-38. [ Links ]
23. Ordóñez N, Castañeda E. Serotipificación de aislamientos clínicos y del medio ambiente de Cryptococcus neoformans en Colombia. Biomédica. 1994;14:131-9. [ Links ]
24. Viviani MA, Esposto MC, Cogliati M, Montagna MT, Wickes BL. Isolation of a Cryptococcus neoformans serotype A MATa strain from the Italian environment. Med Mycol. 2001;41:383-6. [ Links ]














