Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Biomédica
versão impressa ISSN 0120-4157versão On-line ISSN 2590-7379
Biomédica v.29 n.4 Bogotá out./dez. 2009
ARTÍCULO ORIGINAL
1 Grupo de Micobacterias, Instituto Nacional de Salud, Bogotá D.C., Colombia
2 Centro Colombiano de Investigación en Tuberculosis-CCITB, Bogotá D.C., Colombia
3 Maestría en Ciencias Biológicas, Facultad de Ciencias, Pontificia Universidad Javeriana, Bogotá D.C., Colombia
4 Maestría en Biología Molecular y Biotecnología, Facultad de Ciencias Básicas, Universidad de Pamplona, Pamplona, Colombia
5Escuela de Bacteriología y Laboratorio Clínico, Universidad Industrial de Santander, Bucaramanga, Colombia
Recibido: 06/11/08; aceptado:30/04/09
Introduction. Manipulating Mycobacterium tuberculosis clinical specimens and cultures represents a risk factor for laboratory personnel. One of the processes that requires high concentrations of microorganisms is DNA extraction for molecular procedures. Pulmonary tuberculosis cases have occurred among professionals in charge of molecular procedures that require manipulation of massive quantities of microorganisms. This has prompted research studies on biosafety aspects of extraction protocols; however, as yet, no consensus has been reached regarding risks associated with the process.
Objective. The biosafety was evaluated for the DNA extraction protocol of van Soolingen, et al. 2002 by determining M. tuberculosis viability at each process stage.
Materials and methods. Eight hundred eighty cultures were grown from 220 M. tuberculosis clinical isolates that had been processed through the first three DNA extraction stages. Molecular identifications of positive cultures used a PCR isolation of a fragment of the heat shock protein PRA-hsp65 and examination of its restriction enzyme profile (spoligotyping).
Results. Growth was seen in one culture with one of the procedures used. The molecular characterization did not correspond to the initially analyzed isolate, and therefore was deduced to be the product of a cross-contamination.
Conclusion. The DNA extraction protocol, as described by van Soolingen, et al. 2002 and as implemented at the Instituto Nacional de Salud, was established to be safe for laboratory personnel as well as for the environment.
Key words: Mycobacterium tuberculosis, exposure to biological agents, DNA, laboratory techniques, DNA procedures.
Evaluación de la bioseguridad del protocolo de extracción de ADN para especies del complejo Mycobacterium tuberculosis implementado en el Instituto Nacional de Salud
Introducción. El trabajo con Mycobacterium tuberculosis se considera un factor de riesgo para el personal de laboratorio que manipula especímenes clínicos y cultivos. Uno de los procesos que requiere de una alta concentración de microorganismos es la extracción de ADN para realizar metodologías moleculares. Se han reportado casos de tuberculosis pulmonar en profesionales que realizan procedimientos moleculares en los que se requiere previa manipulación del microorganismo en masa, lo cual ha motivado la investigación sobre la bioseguridad del protocolo de extracción, sin que a la fecha haya consenso sobre los riesgos del proceso.
Objetivo. Evaluar la bioseguridad del protocolo de extracción de ADN reportado por van Soolingen et al., 2002, mediante la determinación de la viabilidad de M. tuberculosis en cada etapa del proceso.
Materiales y métodos. Se realizaron 880 cultivos a partir de 220 aislamientos clínicos de M. tuberculosis que se procesaron para las tres primeras fases de extracción de ADN. A los cultivos positivos se les realizó identificación molecular por PRA hsp65 y caracterización por spoligotyping.
Resultados. Se obtuvo crecimiento en uno de los procedimientos realizados. Por caracterización molecular, se determinó que no correspondió al aislamiento analizado originalmente, sino que fue producto de contaminación cruzada.
Conclusión. Se determinó que el protocolo de extracción de ADN descrito por van Soolingen et al. (2002) e implementado en el Instituto Nacional de Salud de Colombia, es seguro para el personal de laboratorio y el medio ambiente.
Palabras claves: Mycobacterium tuberculosis, exposición a agentes biológicos, ADN, técnicas y procedimientos de laboratorio.
Mycobacterium tuberculosis is the etiological agent of tuberculosis, a disease affecting 9.2 million people annually with a 20% mortality rate (1). This world-wide health problem has been excerbated by increasing numbers of susceptible populations due to spread of HIV, malnourishment, immunosuppressant environmental conditions, and M. tuberculosis multidrug or extensively drug resistant isolates often associated with HIV infections (2,3). Key issues in tuberculosis management to stop chains of transmission are quick diagnosis, adequate treatment and location of contacts (4).
As diagnosis is one of the most important aspects, effective molecular methods have been developed to characterize not only the M. tuberculosis complex, but other clinically important non tuberculous mycobacteria complexes; Additional methods can also differentiate clinical isolates based on M. tuberculosis genomic DNA polymorphisms (5-8). In addition to providing a quick identification, these methods decrease the risk of infection during the test procedure--in contrast to conventional methods. Nonetheless, during the past decade, pulmonary tuberculosis cases have been reported among laboratory personnel in charge of molecular procedures aimed at mycobacteria identification (9, Bemer-Melchior P, Gouzerh M, Drugeon H. Transmission of Mycobacterium tuberculosis in a mycobacteriology laboratory. In: Program and Abstracts of the 5th International Conference on the Prevention of Infection; 1998. p. 113.). These cases have caused serious concern among researchers in the field. Recent studies have shown that DNA extraction procedures using M. tuberculosis cultures do not ensure total microorganism inactivation and, therefore, are not completely safe (10,11). Some of these molecular procedures require large quantities of DNA and require the manipulation of high concentrations of living bacilli during the first stages of extraction processes (12). This increases the probability that some bacteria may remain, since heat inactivation may not be completely effective in a highly concentrated mass of microorganisms.
In molecular procedures such as RFLP (restriction length fragment polymorphism), spoligotyping and MIRU (mycobacterium interspersed repetitive units), the DNA extraction protocol of choice is the one described by Van Soolingen et al. 2002,and modified from the Van Embden et al. 1993 protocol. This consists of the following steps: (1) removing the bacillar mass from liquid or solid cultures, (2) heat inactivation of the mycobacteria, (3) enzyme lysis, and (4) extraction of the DNA. The protocol describes each step on the basis of biosafety (BS) levels, with step 1 held at BS-3, and the remaining steps at BS-2 (7,13).
Potential dangers during M. tuberculosis culture manipulation are well known (10,11,14,15). However, few reports have evaluated the biosafety of the protocols, especially during the initial stages (16). Furthermore, authors disagree about what constitutes biosafety during heat inactivation and enzyme lysis steps, since viable microorganisms have been reported during the enzyme lysis stage even after lysozyme and proteinase K treatments (10,14,16). In contrast, other studies have demonstrated that a 20-minute heat inactivation at 80 °C for M. tuberculosis concentrated suspensions provides sufficient level of safety for laboratory personnel (17).
Clearly, the safety of the DNA extraction protocol must be evaluated in each laboratory to establish if the tuberculosis bacilli become completely inviable during the process. If that is not the case, procedures must be implemented to inactivate completely the bacillar suspension without sacrificing DNA quality (17). Several options have been proposed, such as increasing inactivation temperature or carrying out the entire extraction process under level BS-3 conditions (11). The latter considerably reduces infection risks without affecting DNA quality; its use to specialized laboratories. This will directly affect research development in countries with high tuberculosis incidence rates (18). More recent studies have proposed several key measures regarding the M. tuberculosis inactivation process, relating to suspension concentrations and complete immersion of vials in the inactivation medium (generally sterile water) (16,19).
The current study evaluated biosafety in the DNA extraction protocol described by Van Soolingen et al. 2002 (13) by determining M. tuberculosis viability during the first process stages to establish if biosafety measures implemented were effective and guaranteed biosafety for laboratory personnel in the Grupo de Micobacterias from the Instituto Nacional de Salud. This was done while monitoring for environment protection and risk to DNA quality.
Materials and methods
Source of isolates
Two hundred twenty M. tuberculosis clinical isolates were processed between March 2007 and March 2008 and stored at the mycobacteria bank of the Instituto Nacional de Salud.
A total of 880 cultures were obtained from the 220 M. tuberculosis clinical isolates. Each isolate was reconstituted from cryovials stored at -70 °C; they were cultured in Löwenstein Jensen (LJ) medium and incubated at 37 ºC for 15 days. From each culture in exponential phase, a bacillar mass of the isolate was obtained by scraping and then resuspending in two 400 ml vials with TE 1X buffer; these were then inactivated in water bath at 80 °C for 20 minutes; 200 ml of the contents of one of the vials (vial 1) were cultured. Lysozyme (10mg/ml) was then added to both vials, 25 ml to vial 1 and 50 ml to vial 2 (half of the enzyme was added to vial 1 to keep the proportion after having cultured part of the contents); these were then incubated with agitation at 37 °C for one night. After the incubation period, the remaining 200 ml from vial 1 were cultured. Next, 80 ml (7:1) of the SDS mixture 10% / proteinase K from (10mg/ml) were added to vial 2 and incubated in a water bath at 65 °C for 10 minutes; from this solution 200 ml were cultured. Then 50 ml of 5M NaCl and 50 ml of CTAB/NaCl were added to the remaining solution in the vial and incubated for 10 minutes at 65 °C. Next, 200 ml from vial 2 were cultured. The cultures were monitored weekly to observe colony growth, contamination, liquefaction or changes in color. Media showing bacterial growth compatible with a mycobacteria phenotype were processed for molecular identification (figura 1).
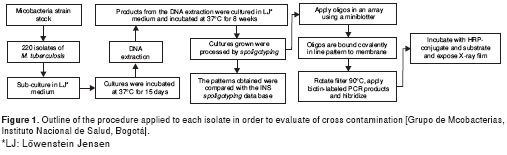
Molecular identification and characterization
Identification of isolates obtained during the protocol evaluation process was done by restriction analysis of hsp65 gene to determine if positive cultures belonged to Mycobacterium genus. Spoligotyping was used to identify M. tuberculosis complex species and determine the genetic pattern of each (6,8).
Results
No growth was observed in any of the cultures during the inactivation stag--80°C and high humidity for 20 minutes. However, during cellular lysis with lysozyme, one tube of the cultures developed a light yellow-colored wrinkled colony after four weeks of incubation. No growth was observed in cultures during proteinase K and CTAB stages.
The positive culture obtained during the cellular lysis stage was subjected to hsp65 gene restriction analysis and was determined to be a member of the M. tuberculosis complex. Later, by spoligotyping, it was identified as M. tuberculosis with a 7777777607760771 genetic pattern, and it was labeled A2 (isolate 2). The pattern obtained for this isolate was compared with the pattern on record in the Grupo de Micobacterias from the Instituto Nacional de Salud spoligotype data base for the same isolate labeled as A1 (isolate 1 or original isolate). This pattern comparison determine that the isolate obtained during the test procedures did not correspond to the pattern of the original mycobacterial strain A1 and A2 provided different molecular patterns, as represented in figura 2.
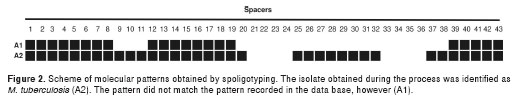
The genetic pattern analysis of the M. tuberculosis isolate determined that the genetic pattern did not correspond to the one recorded in the Instituto Nacional de Salud database for the same isolate. The possibility was discarded that bacilli remained viable through the initial stages of the DNA extraction protocol. The M. tuberculosis genetic pattern was then compared with several clinical isolates processed in the biosafety chamber for other molecular tests the same day. Homology was found with the pattern shown by one of those isolates (figura 2). We concluded that contamination occurred during the procedure, a not uncommon occurrence in laboratories that deal with large amounts of isolates for phenotypic or molecular characterization (20-24). The method used for the identification phase of the current study demonstrated the importance of using spoligotyping; in this case, it permitted clear recognition of cross contamination and confirmed that the protocol used during the extraction process was safe for laboratory personnel. Phenotypic methodologies can reveal the presence of bacilli, but cannot track the molecular identity of the strain after the early stages of DNA extraction.
Although the current study confirmed data obtained by other authors, it is pertinent to mention that this research was conducted primarily to assess the biosafety of the extraction process at the Grupo de Micobacterias at the Instituto Nacional de Salud--the presence of cross-contamination during the process was a fortuitous discovery. No evidence of cross contamination in this laboratory had previously been indicated, possibly due to the lack of warning by other studies in this field. This case recommends systematic monitoring to assess the rate of cross-contamination at each stage of laboratory processing, and setting a maximum rate of contamination of <1%.
The efficiency of M. tuberculosis complex species inactivitation and lysis processes has been carefully studied under the extraction protocol described herein (16,17); however, no consensus has been with regard to its safety for laboratory professionals involved. Differing opinions among authors ranges from those who state that the procedure is only minimally safe to those who maintain that the process is safe with biosafety conditions lower than BS-3 without risk of acquiring the disease (10,11,16-18).
For example, Zwadik et al. 1994 expressed concern about the efficiency of inactiviting mycobacteria by heat, and they reported the occurrence of living microorganisms after 30 min at 95°C. Therefore, they proposed that mycobacteria be inactivated by exposure to humid heat at 100 oC (14). Similarly, Bemer-Melchior et al. 1999 found viable mycobacteria even after the lysozyme and proteinase K treatment and highlighted the Zwadyk et al. recommendations; however, they urged caution with respect to monitoring inactivation times to avoid DNA denaturalization (10). In contrast to these findings, Somerville et al. 2005, using the Van Embden et al. 1993 protocol, carried out a viability study during the DNA extraction procedure, concluded that the protocol was not safe, and proposed that the entire process be confined in biosafety chambers (11). However, the current study has indicated that under the Instituto Nacional de Salud laboratory conditions, the Van Soolingen et al. 2002 extraction protocol ensures M. tuberculosis inactivation from the first stage. This supported the 2002 findings of Doig et al. (17).
In addition to the bacillary load, one of the variables that may influence on bacillus inactivation is the immersion depth of the vial in water during the 20-min period at 85 °C (17,19). This variable was better controlled by submerging the vials almost completely to ensure inactivation of all the bacillar contents and as an important step in ensuring the protocol biosafety.
Under the conditions of the current study, no viable microorganisms were obtained in heat inactivation and enzyme lysis stages. The procedure as described proved safe for laboratory personnel and for the environment as well as ensuring that sufficient DNA was obtained for the molecular procedures required. This was verified by gel quantification and observation, checking at the same time to be sure that no DNA denaturalization occured during heat inactivation or any of the other procedure steps.
Because discrepancies in these procedures are a consequence of specific laboratory conditions (making difficult the standardizing of experimental protocols), the conclusions recommend that molecular biology laboratories reevaluate the biosafety protocols and work plan design to control variables that may have impact on the bacterial inactivation efficacy. These variables may reside in the composition of the inactivation vials, inactivation temperature and its control, bacillary mass concentration, inactivation time, and distribution of the working areas.
The authors state that they have no conflict of interests.
Instituto Nacional de Salud, Centro Colombiano de Investigación en Tuberculosis- CCITB y Colciencias (código 34261817270).
Correspondencia:
Wellman Ribón, Facultad de Salud, Universidad Industrial de Santander, Bucaramanga, Colombia, Carrera 32 N° 29-31, edificio Roberto Serpa, oficina 106, Bucaramanga, Colombia wellmanribon@yahoo.es
1. World Health Organization. Global tuberculosis control: Surveillance, lanning, nancing. WHO Report 2008 (WHO/HTM/TB/2008.393). Geneva: World Health Organization; 2008. [ Links ]
2. Bentwich Z, Maartens G, Torten D, Lal A, Lal R. Concurrent infections and HIV pathogenesis. AIDS. 2000;14:2071-81. [ Links ]
3. World Health Organization. Guidelines for the programmatic management of drug-resistant tuberculosis. WHO/HTM/TB/2006.361. Geneva: World Health Organization; 2006. [ Links ]
4. Broekmans J. Control strategies and programme management. In: Porter JD, McAdam PW, editors. Tuberculosis, back to the future. New York, N.Y: John Wiley & Sons Inc.; 1994. p. 171-92. [ Links ]
5. Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762-71. [ Links ]
6. Devallois A, Goh K, Rastogi N. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 myco-bacterial species. J Clin Microbiol. 1997;35:2969-73. [ Links ]
7. Van Embden J, Cave M, Crawford J, Dale J, Eisenach K, Gicquel B. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: Recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406-9. [ Links ]
8. Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907-14. [ Links ]
9. Menzies D, Fanning A, Yuan L, Fitzgerald M. tuberculosis among health care workers. N Engl J Med. 1995;332:92-8. [ Links ]
10. Bemer-Melchior P, Drugeon H. Inactivation of Mycobacterium tuberculosis for DNA typing analysis. J Clin Microbiol. 1999;37:2350-1. [ Links ]
11. Somerville W, Thibert L, Schwartzman K, Behr M. Extraction of Mycobacterium tuberculosis DNA: a question of containment. J Clin Microbiol. 2005;43:2996-7. [ Links ]
12. Van Soolingen D, Hermans P, de Haas P, Soll D, van Embden J. The occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578-86. [ Links ]
13. Van Soolingen D, de Hass P, Kremer K. Restriction fragment length polymorphism (RFLP) typing of Mycobacteria. Bilthoven: National Institute of Public Health and the Environment; 2002. [ Links ]
14. Zwadyk P Jr, Down J, Myers N, Dey M. Rendering of mycobacteria safe for molecular diagnostic studies and development of a lysis method for strand displacement amplification and PCR. J Clin Microbiol. 1994;32:2140-6. [ Links ]
15. Collins C, Kennedy D. Laboratory-acquired infections: History, incidence, causes and prevention. 4th edition. Oxford: Butterworth-Heinemann; 1999. [ Links ]
16. Blackwood K, Burdz T, Turenne C, Sharma M, Kabani A, Wolfe J. Viability testing of material derived from Mycobacterium tuberculosis prior to removal from a containment level-III laboratory as a part of a laboratory risk assessment program. BMC Infect Dis. 2005;5:1-7. [ Links ]
17. Doig C, Seagar A, Watt B, Forbes K. The efficacy of the heat killing of Mycobacterium tuberculosis. J Clin Pathol. 2002;55:778-9. [ Links ]
18. Warren R, Kock M, Engelke E, Myburgh R, Van Pittius N, Victor T, et al. Safe Mycobacterium tuberculosis DNA extraction method that does not compromise integrity. J Clin Microbiol. 2006;44:254-6. [ Links ]
19. Djelouagji Z, Drancourt M. Inactivation of cultured Mycobacterium tuberculosis organisms prior to DNA extraction. J Clin Microbiol. 2006;44:1594-5. [ Links ]
20. Glynn J, Yates M, Crampin A. DNA fingerprint changes in tuberculosis: reinfection, evolution, or laboratory error? J Infect Dis. 2004;190:1158-66. [ Links ]
21. Small P, McClenny N, Singh S, Schoolnik G, Tompkins L, Mickelsen P. Molecular strain typing of Mycobacterium tuberculosis to confirm cross-contamination in the mycobacteriology laboratory and modification of procedures to minimize occurrence of false-positive cultures. J Clin Microbiol. 1993;31:1677-82. [ Links ]
22. Wurtz R, Demarais P, Trainor W. Specimen contamination in mycobacteriology laboratory detected by pseudo-outbreak of multidrug-resistant tuberculosis: analysis by routine epidemiology and confirmation by molecular technique. J Clin Microbiol. 1996;34:1017-9. [ Links ]
23. Burman W, Stone B, Reves R. The incidence of false-positive cultures for Mycobacterium tuberculosis. Am J Respir Crit Care Med. 1997;155:321-6. [ Links ]
24. Bhattacharya M, Dietrich S, Mosher L. Cross contamination of specimens with Mycobacterium tuberculosis: clinical significance, causes, and prevention. Am J Clin Pathol. 1998;109:324-30. [ Links ]














