Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Biomédica
Print version ISSN 0120-4157
Biomédica vol.32 no.2 Bogotá Apr./June 2012
ARTÍCULO ORIGINAL
1Laboratorio de Genética Molecular Bacteriana, Universidad El Bosque, Bogotá, D.C., Colombia
2Infection, Immunity and Innovation-I3 Institute, Faculty of Science, University of Technology, Sidney, Australia; Institución donde se realizó el estudio: Universidad El Bosque
Author contributions:
Javier Antonio Escobar and Natasha Vanegas conceived and designed the experiments.
Ricaurte Alejandro Márquez, Bibiana Chavarro, Ingrid Tatiana Gómez, Betsy Esperanza Castro and Martha Johanna Murillo performed the experiments.
All authors analyzed the data and wrote the paper.
Recibido: 14/07/11; aceptado:13/02/12
Introduction. Community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) infections are found with increasing the frequency, both in healthy individuals in the community and in hospitalized patients. In Colombia and the Andean region, CA-MRSA isolates have a genetic background that is related to the pandemic USA300 clone.
Objective. Two molecular methods are designed and standardized for the rapid differentiation of Colombian community-acquired and hospital-acquired methicillin-resistant Staphylococcus aureus (HA-MRSA) isolates.
Materials and methods. Two molecular methods were standardized for the identification of CA-MRSA isolates. The first method was based on the differential digestion of the carbamate kinase (arcC)and guanylate kinase (gmk) genes in the sequences type 5 (ST5) in the HA-MRSA isolates and 8 (ST8) in the CA-MRSA isolates. The second method was based on the PCR amplification of 5 specific virulence factors found in CA-MRSA and HA-MRSA isolates. The specificity and precision of each method were evaluated using 237 clinical MRSA isolates.
Results. The first method identified 100% and 93.2% of the CA-MRSA and HA-MRSA isolates, respectively. The second method also correctly identified the two isolates types (CA-MRSA and HA-MRSA).
Conclusions. These two methods are a convenient alternative for the rapid identification of the CA-MRSA isolates, compared with other techniques such as pulsed field gel electrophoresis and multilocus sequence typing, which are time-consuming and more expensive.
Key words: Methicillin-resistant Staphylococcus aureus, community-acquired infections, bacterial typing techniques, multilocus sequence typing, enterotoxins.
Diseño de dos metodologías moleculares para la rápida identificación de aislamientos de Staphylococcus aureus resistente a meticilina asociados a la comunidad en Colombia
Introducción. Los aislamientos de Staphylococcus aureus resistente a la meticilina asociado a la comunidad (SARM-AC), están aumentando la frecuencia de infecciones en personas sanas de la comunidad y en pacientes hospitalizados. En Colombia y en la región andina estos aislamientos tienen un componente genético relacionado con el clon pandémico USA300.
Objetivo. Diseñar y estandarizar dos metodologías para la diferenciación rápida de aislamientos colombianos de S. aureus resistente a la meticilina asociado a la comunidad de los asociados al hospital (SARM-AH).
Materiales y métodos. Se estandarizaron dos metodologías moleculares para la identificación de aislamientos de S. aureus resistente a la meticilina asociado a la comunidad. La primera se basa en la digestión diferencial con tres enzimas de restricción de los genes cinasa de carbamato (arcC)y cinasa de guanilato (gmk)para los tipos de secuencia 5 (ST5) y 8 (ST8), correspondientes a aislamientos de S. aureus resistente a la meticilina asociado al hospital y asociado a la comunidad, respectivamente. La segunda se basa en la amplificación por reacción en cadena de la polimerasa de cinco factores de virulencia que se encuentran de manera diferencial en estos aislamientos. Las dos metodologías fueron validadas en 237 aislamientos clínicos de S. aureus resistente a la meticilina.
Resultados. Con la primera metodología se identificaron el 100 % y 93,2 % de los aislamientos de S. aureus resistente a la meticilina asociado a la comunidad y asociado al hospital, respectivamente. Con la segunda metodología se identificaron correctamente los dos tipos de aislamientos.
Conclusiones. Estas dos metodologías son una buena alternativa en términos de ahorro en tiempo y dinero comparadas con otras técnicas, como la electroforesis en campo pulsado y la tipificación de secuencias multilocus para la rápida identificación de aislamientos de S. aureus resistente a la meticilina asociado a la comunidad en Colombia.
Palabras clave:Staphylococcus aureus resistente a meticilina, infecciones comunitarias adquiridas, técnicas de tipificación bacteriana, tipificación de secuencias multilocus, sequence typing, enterotoxinas.
The first infections due to community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) were reported in Australia in 1990 (1). Since then the number of reported cases has increased around the world, demonstrating a high capacity for infection and dissemination (2). In addition, the CA-MRSA isolates have begun to cause infections in hospitalized patients and to replace the clones of hospital-acquired methicillin-resistant Staphylococcus aureus (HA-MRSA) (3,4). These isolates differ from the HA-MRSA forms in that they are more virulent and possess genetic components that cause infections even in healthy persons without apparent risk factors or prior contact in a hospital environment (2,5,6). Even though HA-MRSA has been a clinical concern for a long time, the infections caused by CA-MRSA have become the focal point for clinical and researchconcern.
Methicillin-resistant Staphylococcus aureus (MRSA) presents a conserved clonal structure with a reduced number of clones that have the capacity for global dissemination. Five CA-MRSA pandemic clones that have been described based on their sequence type (ST), as determined by multilocus sequence typing (MLST): European clone (ST80); Oceanic clone (ST30); North-West Pacific clone (ST59); USA300 (ST8) and USA400 (ST1)(2). In South America, the appearance and dissemination of at least three of the CA-MRSA clones have been reported, but with different genetic characteristics. In Brazil and Uruguay the most prevalent CA-MRSA clone possesses a type of staphylococcal cassette chromosome mec (SCCmec) IVc, the Panton-Valentine leukocidin (PVL) toxin and ST30. These are genetically related to the CA-MRSA clone in Australia and Oceania. In Argentina, a CA-MRSA clone has been reported with a SCCmec IVa, positive PVL, ST5 and with no apparent genetic relation to one of the CA-MRSA pandemic clones (7,8). In contrast, the CA-MRSA clone that circulates in the Andean region (Colombia, Ecuador, Peru and Venezuela) is related to the USA300 clone, possesses an ST8, a spa type t008, has PVL and a SCCmec IVc (9,10).
In Colombia, two predominant HA-MRSA clones have been found--the Chilean/Cordobés and the Pediatric clones. The first has SCCmec type Iand the second has a SCCmec type IV. These clones do not possess the LukF-PV/LukS-PV genes, and both have an ST5 (allele combination 1, 4, 1, 4, 12, 1, 10) and belong to the clonal complex 5. In contrast, the Colombian CA-MRSA isolates are related to the USA300 clone and have an ST8 (allele combination 3, 3, 1, 1, 4, 4, 3).
In microbiological terms, CA-MRSA and HA-MRSA isolates are very difficult to differentiate. Techniques such as pulsed field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) must be used. PFGE is based on the digestion of the chromosomal bacterial DNA with a restriction enzyme (SmaI in the case of S. aureus). The DNA fragments obtained are separated by PFGE, generating a banding pattern (pulsotype) (11). The second method, MLST, identifies the allelic variation, at the sequence level, of 7 standard housekeeping genes chosen for S. aureus--carbamate kinase (arcC), shikimate kinase (aroE), glycerol kinase (glpF), guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi) and acetyl coenzyme A acetyltransferase (yqiL).Each combination of the 7 alleles provides a specific sequence type in accordance with the MLST database (available at www.mlst.net). Isolates with related STs determine a clonal complex (CC). The use of these techniques has allowed the differentiation of the pandemic MRSA clones (12-15).
PFGE and MLST are expensive techniques that are time-consuming and require a staff trained in the use of specialized facilities and equipment. Additionally, analysis of results and the comparison of the pulsotypes are not straightforward tasks. For these reasons, the development of methods has become important to allow the rapid identification of CA-MRSA isolates and provide improved treatment of the infections caused by these microorganisms.
Therefore, the objective was to design and standardize two methods for the rapid identification of Colombian CA-MRSA isolates related to the USA300 clone.
Materials and methods
Staphylococcus aureus isolates
The MRSA clinical isolates were derived from strains banked at the Bacterial Molecular Genetic Laboratory of the Universidad El Bosque in Bogotá, Colombia. The reference strains used were as follows: USA300-0114 (ST8-MRSA-IVa), CHL93, corresponding to the Chilean/Cordóbes clone (ST5-MRSA-I), and HDE3, corresponding to the Pediatric clone (ST5-MRSA-IV). Two hundred and thirty seven MRSA clinical isolates were analyzed--197 isolates (153 CA-MRSA and 44 HA-MRSA) were previously classified as CA-MRSA and HA-MRSA. Genetic relationships established in circulating clones by means of PFGE and MLST and microbiological factors such as minimal inhibitory concentration (MIC) for oxacillin, gentamicin, clindamycin, erythromycin, ciprofloxacin, tetracycline, chloramphenicol, rifampicin, trimethoprim-sulfamethoxazole, vancomycin and linezolid. In addition, 40 MRSA isolates were first classified as CA-MRSA and HA-MRSA using the two methodologies standardized, and then the isolates were confirmed by molecular and genetic characterization.
DNA extraction
The isolates were recovered in brain-heart infusion broth (BHI) after incubation for 24 hours at 37°C under aerobic conditions. The DNA was extracted by resuspension of the colony in 30 µL of distilled and deionized water and later boiled at 94°C for 10 min. Finally, it was centrifuged at 5,000 rpm for five min and 5 µL of supernatant solution was used as the DNA template for each reaction.
Determination of the allelic variations and the search for specific restriction enzymes
The sequences for the alleles of the genes arcC, aroE, glpF, gmk, pta, tpi and yqiL for ST5 and ST8 were taken from the MLST website at www.mlst.net. The nucleotide variations for each of the genes and the search for differentiating restriction enzymes were determined by means of the Restriction of DNA program (16). These results were confirmed by means of multiple alignment of each allelic pair using the Multalin program, available at www.expasy.org. The cutting sites for the chosen restriction enzymes were confirmed through the programs NEBcutter (17) and Webcutter, available at http://rna.lundberg.gu.se/cutter2/.
Amplification of the genes arcC (carbamate kinase) and gmk (guanylate kinase) and cutting with restriction enzymes
The design of the primers was made using the total sequence of each gene, downloaded as part of the genome of the USA300 strain that was available in GenBank (access number gi87159884) and using the programs PRIMER and PRIMER3 (18,19). The parameters used for the design of the primers were as follows: (1) the size of the product to amplify, (2) size of the products generated after the restriction (easy identification of the alleles), (3) number of additional cuts of each restriction enzyme inside the amplified product, (4) a GC percentage between 40-60%, (5) a maximum variation of 5°C in their annealing temperatures, and (6) a lack of formation of secondary structures. The optimal concentrations of MgCl2, dNTPs, primers and DNA were determined, as were the best thermal conditions, by a trial and error approach. The amplified products of each gene were digested with the selected restriction enzymes at 37°C for 2 hours. Finally, the restriction patterns were visualized in 1.5% agarose gels dyed with ethidium bromide.
Multiple amplification of the genes sek, bsaB, lukF-PV/lukS-PV, sem and seo
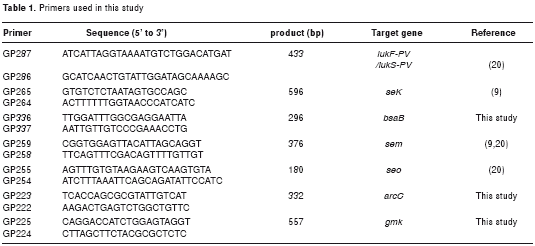
A multiple PCR was standardized for the simultaneous amplification of the genes sek, bsaB, lukF-PV/lukS-PV, sem and seo that codify for the enterotoxin K, bacteriocin B, PVL, enterotoxin M and enterotoxin O, respectively. The primers used for the amplification of these genes are shown in table 1.
Results
First Method: Differential digestion of the carbamate kinase (arcC) and guanylate kinase (gmk) genes in the sequences type 5 (ST5) and 8 (ST8)
The sequence types ST5 and ST8 have different alleles in 6 of the 7 genes used for the MLST (arcC, aroE, gmk, pta, tpi and yqiL). The allele 1 of the gene glpF is the same for these two types of ST. The nucleotide variations of each allelic pair were determined for each of the 6 genes in the two ST types by means of multiple alignments. In total, 17 variations were found--3 in the arcC gene, 2 in the aroE gene, 5 in the gmk gene, 1 in the pta gene, 2 in the tpi gene and 4 in the yqiL gene (table 2). For each allelic pair, sequences of 23 nucleotides were taken that included each variation (10 nucleotides on each side of the variation); then the differential cutting restriction enzymes were selected by means of the “Restriction of DNA”, Webcutter 2.0 and NEBcutter V2.0 programs. Appropriate restriction enzymes found were for only some variations of the arcC and gmk genes. For the arcC gene, the HinfI enzyme (GANTC cutting and recognition sequence) cut allele 1 of ST5 in the position 198, but it did not cut allele 3 of ST8 for the nucleotide variation C198T. For the gmk gene, two restriction enzymes cut differently in three of the five variants. The HhaI enzyme (GCGC cutting and recognition sequence) cut only allele 1 for ST8 in the position 286 but it did not cut allele 4 of ST5 for the nucleotide variation T286C. The CaiI enzyme (with CAGNNNCTG recognition sequence) cut allele 1 of ST8 but not allele 4 of ST5 for the consecutive variations C357T and A358G (table 2).
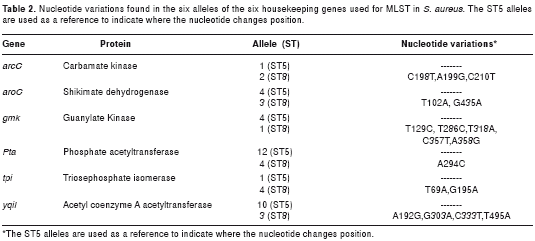
The arcC gene has a total size of 942 bp; the designed primers amplify an expected product of 332 bp and include the variation C198T. The gmk gene has total size of 624 bp. For the primer design, the amplified product was expected to include all of the 3 variants, T286C, C357T and A358G. The expected size was 557 bp (table 1).
After a standardization process, the optimal concentrations of the reagents for the amplification of the arcC and gmk genes were as follows: 2.5 mM of MgCl2, 200 µM of dNTPs, 200 nM of each primer (forward and reverse) and 2 units of Taq Polymerase. The two genes were amplified using the same temperature profile, which consisted of: an initial cycle of 94°C for 5 min, followed by 30 cycles of 94°C for 30 sec, 50°C for 30 sec and 72°C for 1 min; and a final extension cycle of 72°C for 7 min in a final volume of 20 µL. Later, 10 µL of the PCR product, without previous purification, was incubated at 37°C with each of the respective restriction enzymes for 2 hours. For the restriction of the arcC gene, 7 units of the HinfI enzyme were used, and the fragments expected in accordance with the ST were as follows: 332 bp for an isolate with allele 3 (ST8), the amplified product size (uncut by the restriction enzyme), and two products of 245 bp and 87 bp for allele 1 (ST5) (figure 1). For the restriction of the gmk gene, 5 units of the CaiI or HhaI enzymes were used. The expected fragments with the CaiI enzyme were 557 bp for the allele 4 (ST5), uncut PCR product, and two products of 390 bp and 167 bp for allele 1 (ST8). In the case of the HhaI enzyme, this cuts the two alleles of the gmk gene, but produces a different number of fragments--for allele 4 of ST5 it cuts once and produces two fragments of 429 bp and 124 bp;, for allele 1 of ST8 it cuts twice and produces three fragments of 323 bp, 124 bp and 105 bp (figure 1).
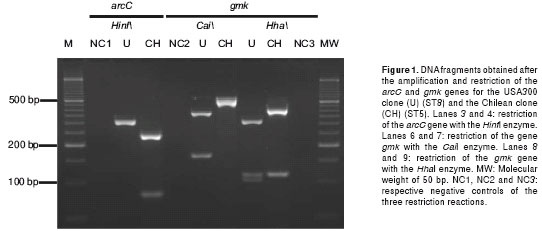
The restriction of the arcC gene with the HinfI enzyme in the Chilean clone (ST5) produced two fragments of 245 bp and 87 bp. In the case of the USA300 clone (ST8), a fragment of 332 bp was obtained that corresponded to the size of the amplified product, i.e., no cutting occurred with this enzyme. The restriction of the gmk gene with the CaiI enzyme in the USA300 clone (ST8) produced two fragments of 340 bp and 112 bp. For the Chilean clone (ST5) a fragment of 557 bp was obtained, indicating no activity of the restriction enzyme. The restriction of the gmk gene with the HhaI enzyme in the Chilean clone (ST5) produced two fragments of 428 bp and 129 bp, and for the USA300 clone (ST8) three fragments of 323 bp, 129 bp and 105 bp (figure 1). The results obtained for the Pediatric clone HDE3 (ST5) were the same as those found for the Chilean clone.
From these results, the following deductions were made. (1) The HinfI enzyme cut allele 1 of the arcC gene in the ST5 isolates, but it did not cut allele 3 of the ST8 isolates. (2) The CaiI enzyme cut allele 1 of the gmk gene present in the ST8 isolates, but it did not cut allele 4 of ST5 isolates. (3) In the case of the HhaI enzyme, it cut the two alleles of the gmk gene, but with a different frequency and generated fragments with different sizes. (4) Allele 1, present in the isolates with ST8, was cut at two sites and generated three products. (5) For allele 4, present in ST5 isolates, HhaI cut in only one site and generated two products, one with a greater size (428 bp) than that expected for allele 1 (323 bp).
Second method: PCR amplification of the 5 specific virulence factors
Chavarro et al. (2009) carried out a study on the frequency of 24 virulence factors in 270 Colombian MRSA isolates (86 CA-MRSA isolates and 184 HA-MRSA isolates) (9). The sek, seq and lukF-PV/lukS-PV genes were found only in CA-MRSA isolates with a frequency of 80%, 82% and 92%, respectively. The seg, sei, sem, sen and seo genes, that make up the enterotoxin genomic cluster (egc) were found only in the HA-MRSA isolates with a frequency of 85%. Recently Álvarez et al. analyzed 153 MRSA isolates recovered from pediatric infections and found the sek, seq and lukF-PV/lukS-PV genes only in the CA-MRSA isolates, with frequencies of 71%, 71% and 100%, respectively. The egc genes were found in all of the HA-MRSA isolates. In addition, they found that 90% of the CA-MRSA isolates had the bsaB gene that codifies for a bacteriocin, whereas this gene was not found in the HA-MRSA isolates (unpublished data).
Using these results, primers were specifically designed for the simultaneous amplification of the lukF-PV/lukS-PV, sek, bsaB, sem and seo genes. The first three genes act as specific genetic markers for Colombian CA-MRSA isolates and the remaining two for Colombian HA-MRSA isolates (figure 2). The primer sequences and the size of the expected amplification products are detailed in table 1.
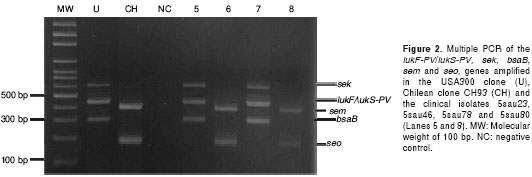
The optimal concentrations of the reagents found for the simultaneous amplification of the 5 genes were as follows: 2.5 mM of MgCl2, 200 µM of dNTPs, 200 nM of each primer and 1 unit of Taq polymerase. The optimal profile of temperatures was: an initial 95°C for 5 min, followed by 30 cycles of 95°C for 45 sec, then 54°C for 45 sec and 72°C for 2 min, with a final extension of 72°C for 7 min (figure 2).
Validation of the two methods
The methods were validated evaluating 197 and 40 MRSA clinical isolates, with and without micro-biological, genetic and molecular characterization, respectively. Among the 197 MRSA isolates previously characterized, 153 (77.6%) were classified as CA-MRSA and 44 (22.3%) were classified as HA-MRSA. With the first method, the 153 CA-MRSA isolates evaluated showed a restriction profile of the genes arcC and gmk expected of CA-MRSA isolates, while of the 44 HA-MRSA isolates analyzed, 41 showed restriction profiles that were characteristic of HA-MRSA isolates and 3 (6.8%) isolates presented profiles with restriction characteristics of CA-MRSA isolates for the HinfI and CaiI enzymes. However, the amplified fragment of the gmk gene was not cut by the HhaI enzyme in those three isolates, an unexpected result for the ST5 and ST8 isolates. This suggests the possibility that these 3 isolates possess a different allele of the gene gmk. To confirm this hypothesis, the allele of this gene will be sequenced for these samples. These results show that the first standardized method had a high specificity (98.5%) and correctly identified all of the CA-MRSA isolates. The second method correctly identified both the CA-MRSA and HA-MRSA isolates (table 3). The 153 CA-MRSA isolates amplified at least one of the three genes (sek, bsaB or lukF-PV/lukS-PV). One hundred thirteen (73.8%) isolates amplified the three genes, 29 (18.9%) only amplified the lukF-PV/lukS-PV gene, 8 (5.2%) amplified the sek and bsaB genes and 3 (2.0%) isolates amplified the lukF-PV/lukS-PV and bsaB genes. All of the HA-MRSA isolates amplified the sem and seo genes.
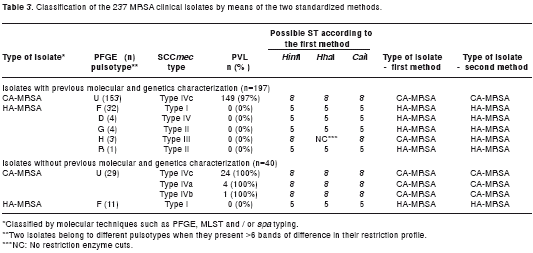
Among the 40 MRSA clinical isolates without previous molecular characterization, 29 (72.5%) were classified as CA-MRSA and 11 (27.5%) were classified as HA-MRSA using the two methods. With the first method, the 29 CA-MRSA isolates evaluated showed a restriction profile of the genes arcC and gmk expected of CA-MRSA isolates, the 11 HA-MRSA isolates analyzed showed restriction profiles that were characteristic of HA-MRSA isolates (100% exactitude). With the second method, the 29 CA-MRSA isolates amplified at least one of the sek, bsaB or lukF-PV/lukS-PV genes and all HA-MRSA isolates amplified the sem and seo genes. The subsequent molecular and genetic characterization showed that all isolates classified CA-MRSA were related to USA300 clone and all isolates classified HA-MRSA were related to Chilean clone. These results confirm that the two methods correctly identified both the CA-MRSA and HA-MRSA isolates (100% exactitude) (table 3).
Discussion
Several international studies have demonstrated that the CA-MRSA isolates are more virulent than the HA-MRSA isolates, due to greater production of virulence factors such as the Panton-Valentine leukocidin (PVL) and the phenol-soluble modulins (PSM), capable of lysing macrophages and neutrophils (5,6). Additionally, the CA-MRSA isolates contains genetic mobile elements that give them greater environmental adaptability--as is the case with the arginine catabolic mobile element (ACME), specifically identified in the USA300 clone (21). Clinically, the CA-MRSA isolates have acquired the capacity not only to cause minor infections in healthy persons, but also to cause severe infections such as pneumonia and necrotizing fasciitis (16). The entry of CA-MRSA to hospitals and their greater virulence indicates an expected increase in patient mortality and morbidity in affected institutions. This underlines the importance of developing methods that allow rapid identification of CA-MRSAs, and hence, more effective treatment, control and eradication of these infections.
Currently, the CA-MRSA and HA-MRSA isolates are differentiated with greater accuracy using robust and expensive techniques like PFGE and MLST. In developing countries such as Colombia and others in the Andean region, faster and cheaper alternative methods are necessary to differentiate between these types of microorganisms in hospitals. Herein,methods have been standardized for the rapid identification of Colombian CA-MRSA isolates; the methods were designed based on isolates from several multi-centre studies (with epidemiological data as well as molecular and genetic characterizations), as well as new isolates without molecular characteristics and later confirmed by PFGE and MLST (4,9,10,22,23).
The first method was based on the detection of polymorphism, by means of restriction enzymes, found in two constitutive genes of S. aureus, which are used in MLST. This method correctly identified the 153 CA-MRSA isolates (100%) and 41 of the 44 HA-MRSA isolates (93.2%). A more detailed analysis of the molecular characteristics of the three HA-MRSA isolates classified as CA-MRSA was conducted. This revealed that these have a SCCmec III and a PFGE pulsotype (named as H, table 3) possibly related to the Brazilian clone. This clone possesses a SCCmec IIIa and an ST239. Its allelic combination is 2-3-1-1-4-4-3, a single locus variant (SLV) from ST8 (3-3-1-1-4-4-3); allele 2 of the arcC gene also possesses the mutation C198T, as found in ST8. For this reason,the restriction enzyme HinfI did not cut the arcC gene in the three isolates, and consequently classified as CA-MRSA. The epidemiological data indicated that the circulation of isolates related to the Brazilian clone is very low, with a frequency of less than 1% (unpublished data). The first method correctly identified all of the “new” 29 CA-MRSA isolates and 11 HA-MRSA isolates (100%).
Various methods have been developed to try to identify CA-MRSA isolates at an international level (24,25). For example, Diep et al. (2003) reported a method that established the genetic relationship between two or more isolates from S. aureus by analyzing the polymorphism of amplified fragments of the 7 genes used for MLST, utilizing restriction enzymes (26). Unfortunately, this method does not allow the easy differentiation between ST5 and ST8 isolates--it generates DNA fragments with very similar size because it uses the primers employed by the MLST.
The second method correctly identified the 197 CA-MRSA and HA-MRSA isolates, including the 3 HA-MRSA isolates possibly related to the Brazilian clone. This method allowed the differentiation between the Colombian CA-MRSA and HA-MRSA isolates by the detection of specific virulence factors for each type of isolate. The fundamental characteristic of these virulence factors are their location on mobile genetic elements, which are acquired differently in the two types of isolates. The sek, bsaB, lukF-PV/lukS-PV genes were selected using results previously obtained by the current study and as reported in other literature (9,10), and the selection supported by a bioinformatics analysis of their location on the genome. In accordance with the genome sequence of the USA300 clone (21), the sek, bsaB, lukF-PV/lukS-PV genes are found on different mobile genetic elements. The first is transported on the pathogenicity island 5 (SaPI5), the second on the genomic island b (vSab) type II and the third on the prophage Sa2usa. The selection of specific virulence factors for the HA-MRSA isolates was made using information from studies previously carried out in our laboratory (the genomes of the Chilean/Cordóbes and Pediatric clones have not yet been sequenced).
These studies verified that approximately 90% of the HA-MRSA isolates contained the egc cluster (9). This cluster has been found on the genomic island b (vSab) type I of the strains N315, Mu50 and Mu3 (27). Fossum et al. (2009) analyzed 821 MRSA isolates and found that each genetic lineage possessed a specific repertoire of enterotoxins. This supports the hypothesis that mobile genetic elements are not inserted in all isolates and are not randomly distributed (28). Miranda et al. (2007) reported the emergence of isolates that were genetically related to the USA800 clone (ST5-SCCmec IV) in Brazil. These were related to the Pediatric clone, with all of the isolates carrying the egc enterotoxin cluster. These observations confirmed that the cluster is widely disseminated in isolates related to the Pediatric clone, not only in Colombia, but also in neighboring countries (29). The second method identified correctly both the CA-MRSA and HA-MRSA isolates (100%), as well as CA-MRSA and HA-MRSA isolates without previous molecular characterization (100%). These results indicate that the two methods may be used for a correct determination a MRSA isolate may be either a CA-MRSA or HA-MRSA isolate.
The two methods standardized in this study allow the identification of the CA-MRSA isolates in a short period of time (approximately 5 hours) and with a lower cost compared to techniques such as PFGE and MLST. Furthermore, even if these methods were based on Colombian CA-MRSA isolates, they have the potential to identify CA-MRSA isolates in the for the entire Andean region (Colombia, Venezuela, Ecuador and Peru), given that the epidemiology of these countries is similar to that of Colombia. The pulsotypes obtained from PFGE show that the majority of the HA-MRSA isolates have a close genetic relation to the Chilean clone; and the CA-MRSA isolates, as in Colombia, are genetically related to the USA300 clone (10,30). These determinations will be of value even if additional studies are required to confirm the STs and whether the STs possess these same specific virulence factors.
The development of these diagnostic tools is an important advance that allows faster and more accurate identifications of the MRSA isolate. Rapid identifications will in turn quickly orientate the medical staff toward a more appropriate empirical antibiotic treatment and assist an adequate therapeutic management of CA-MRSA infections. Finally, without the of knowledge of the genetic and molecular characteristics of the isolates, inappropriate treatments can result in increased resistance of bacterial agents to antibiotics, and, once established, reduces the therapeutic possibilities for treatment.
Acknowledgements
Division of Research of the Universidad El Bosque
Conflicts of Interest
The authors declare that during the conduct of the present study, no conflicts of interest occurred that will have affected the experimental results or the opinions expressed herein.
Funding
This study was financed by the Colombian Administrative Department of Science, Technology and Innovation (COLCIENCIAS) through the project “Standardization of a molecular tool for the differentiation of community acquired (CA-MRSA) and hospital acquired (HA-MRSA) methicillin-resistant Staphylococcusaureus isolates”, code 1308-49326155. It was also supported by the Virginia Gutiérrez de Pineda Young Researchers and Innovators Program and the Division of Research of the Universidad El Bosque.
Correspondencia: Javier Antonio Escobar, Carrera 7B bis Nº 132-11, edificio Rectoría, 2º piso, Universidad El Bosque, Bogotá, D.C., Colombia Teléfonos: (571) 523 4879 y 633 1368, extensión 1179; fax: (571) 625 2030 labgenmolecular@unbosque.edu.co, javiesco21@yahoo.com
References
1. Udo EE, Pearman JW, Grubb WB.Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:97-108. [ Links ]
2. Deleo FR, Otto M, Kreiswirth BN, Chambers HF. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010:375:1557-68. [ Links ]
3. Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787-94. [ Links ]
4. Álvarez CA, Yomayusa N, Leal AL, Moreno J, Méndez-Álvarez S, Ibáñez M, et al.Nosocomial infections caused by community-associated methicillin-resistant Staphylococcus aureus in Colombia. Am J Infect Control. ¿Año?;38:315-8. [ Links ]
5. Diep BA, Palazzolo-Ballance AM, Tattevin P, Basuino L, Braughton KR, Whitney AR, et al. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLoS One. 2008;3:e3198. [ Links ]
6. Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510-4. [ Links ]
7. Sola C, Saka HA, Vindel A, Bocco JL.Emergence and dissemination of a community-associated methicillin-resistant Panton-Valentine leucocidin-positive Staphylococcus aureus clone sharing the sequence type 5 lineage with the most prevalent nosocomial clone in the same region of Argentina. J Clin Microbiol. 2008;46:1826-31. [ Links ]
8. Gardella N, von Specht M, Cuirolo A, Rosato A, Gutkind G, Mollerach M. Community-associated methicillin-resistant Staphylococcus aureus, eastern Argentina. Diagn Microbiol Infect Dis. 2008;62:343-7. [ Links ]
9. Chavarro B. Determinación de la presencia y transcripción de factores de virulencia en aislamientos colombianos de Staphylococcus aureus resistente a meticilina adquirido en la comunidad. Bogotá: Facultad de Medicina, Universidad el Bosque; 2009. [ Links ]
10. Reyes J, Rincon S, Díaz L, Panesso D, Contreras GA, Zurita J, et al.Dissemination of methicillin-resistant Staphylococcus aureus USA300 sequence type 8 lineage in Latin America. Clin Infect Dis. 2009;49:1861-7. [ Links ]
11. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233-9. [ Links ]
12. Tristan A, Ferry T, Durand G, Dauwalder O, Bes M, Lina G, et al. Virulence determinants in community and hospital meticillin-resistant Staphylococcus aureus. J Hosp Infect. 2007;65(Suppl.2):105-9. [ Links ]
13. Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, et al. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol. 2006;44:108-18. [ Links ]
14. Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008;8:747-63. [ Links ]
15. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008-15. [ Links ]
16. Bikandi J, San Millan R, Rementeria A, Garaizar J.In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinformatics. 2004;20:798-9. [ Links ]
17. Vincze T, Posfai J, Roberts RJ. NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003;31:3688-91. [ Links ]
18. Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365-86. [ Links ]
19. Krawetz SA, Womble DD. Design and implementation of an introductory course for computer applications in molecular genetics. A case study. Mol Biotechnol. 2001;17:27-41. [ Links ]
20. Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70:631-41. [ Links ]
21. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731-9. [ Links ]
22. Miller LG, Perdreau-Remington F, Rieg G, Mehdi S, Perlroth J, Bayer AS, et al. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med. 2005;352:1445-53. [ Links ]
23. Álvarez-Olmos MI, Enríquez SP, Pérez-Roth E, Méndez-Álvarez S, Escobar J, Vanegas N, et al.Pediatric cases from Colombia caused by a panton-valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus ST8-SCCmecIVc clone. Pediatr Infect Dis J. 2009;28:935. [ Links ]
24. Bonnstetter KK, Wolter DJ, Tenover FC, McDougal LK, Goering RV. Rapid multiplex PCR assay for identification of USA300 community-associated methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 2007;45:141-6. [ Links ]
25. Strommenger B, Braulke C, Pasemann B, Schmidt C, Witte W. Multiplex PCR for rapid detection of Staphylococcus aureus isolates suspected to represent community-acquired strains. J Clin Microbiol. 2008;46:582-7. [ Links ]
26. Diep BA, Perdreau-Remington F, Sensabaugh GF. Clonal characterization of Staphylococcus aureus by multilocus restriction fragment typing, a rapid screening approach for molecular epidemiology. J Clin Microbiol. 2003;41:4559-64. [ Links ]
27. Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: Polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190:300-10. [ Links ]
28. Fossum AE, Saltyte J, Alm-Kristiansen K, Bukholm G. Exotoxin-encoding gene content in community-acquired and hospital-acquired methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2009;15:1139-45. [ Links ]
29. de Miranda OP, Silva-Carvalho MC, Ribeiro A, Portela F, Cordeiro RP, Caetano N, et al.Emergence in Brazil of methicillin-resistant Staphylococcus aureus isolates carrying SCCmecIV that are related genetically to the USA800 clone. Clin Microbiol Infect. 2007;13:1165-72. [ Links ]
30. Rodríguez-Noriega E, Seas C, Guzmán-Blanco M, Mejía C, Álvarez C, Bavestrello L, et al.Evolution of methicillin-resistant Staphylococcus aureus clones in Latin America. Int J Infect Dis. 2010;14:e560-6. [ Links ]













