Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Biomédica
versão impressa ISSN 0120-4157
Biomédica vol.33 no.1 Bogotá jan./mar. 2013
https://doi.org/10.7705/biomedica.v33i1.643
ARTÍCULO ORIGINAL
doi: http://dx.doi.org/10.7705/biomedica.v33i1.643
1Instituto de Genética, Universidad Nacional de Colombia, Bogotá, D.C., Colombia
2Departamento de Química, Facultad de Ciencias, Universidad Nacional de Colombia, Bogotá, D.C., Colombia
3Departamento de Patología, Facultad de Medicina, Universidad Nacional de Colombia, Bogotá, D.C., Colombia
Authors´ contributions:
Martha Lucía Serrano designed the study and performed the laboratory work.
Juan José Yunis directed the laboratory work.
Both authors analyzed the results and wrote the manuscript.
Recibido: 10/02/12; aceptado:15/08/12
Introduction. Retinoblastoma is a childhood cancer of the retina originated by altered or null retinoblastoma protein (pRb) expression. Genetic alterations in both RB1 alleles in the retinal cells are required for the development of retinoblastoma. In the sporadic form, non-hereditary RB1 gene mutations take place in a single retinoblast cell, and are therefore only present in tumor DNA (somatic mutations). Sporadic retinoblastoma is primarily unilateral, lacks family history and has no risk of transmission to descendants. Genetic tests for detection of RB1 mutation has improved the identification of carriers and facilitated accurate genetic counseling.
Objective. To identify mutations in the RB1 gene in Colombian patients with sporadic retinoblastoma by PCR-SSCP followed by sequence.
Materials and methods. Four patients with sporadic retinoblastoma were analyzed by PCR-SSCP, followed by DNA sequencing to identify variations in the RB1 gene.
Results. We identified five variations in RB1 gene: three new mutations (one germline and two somatic mutations), one new polymorphism and one already reported somatic mutation. Four mutations were found in three patients with unilateral retinoblastoma and one mutation was found in a patient with bilateral retinoblastoma. One of these was a germline mutation in a sporadic unilateral retinoblastoma that was not present in the parents or three siblings analyzed.
Conclusions. Our results emphasize the importance of identifying mutations for genetic counseling and clinical management of sporadic retinoblastoma patients. Description of a new RB1 gene variant is interesting since there have been a small number of polymorphisms reported for this gene.
Keywords: Retinoblastoma/genetics; genes, retinoblastoma; mutations; polymorphism, genetic; Colombia.
doi: http://dx.doi.org/10.7705/biomedica.v33i1.643
Identificación de tres nuevas mutaciones en el gen RB1 en pacientes con retinoblastoma esporádico en Colombia
Introducción. El retinoblastoma es un cáncer pediátrico de la retina originado por la expresión alterada o ausente de la proteína del retinoblastoma (pRb). Se requiere la alteración genética de ambos alelos RB1 en las células de la retina para el desarrollo del retinoblastoma. En la forma esporádica, las mutaciones no hereditarias del gen RB1 ocurren en un solo retinoblasto y están presentes sólo en el ADN del tumor (mutaciones somáticas). El retinoblastoma esporádico es generalmente unilateral, no tiene historia familiar y no tiene riesgo de transmisión a la descendencia. Las pruebas genéticas para la detección de mutaciones en RB1 han mejorado la identificación de portadores y han facilitado la precisión de la asesoría genética.
Objetivo. Detectar mutaciones en el gen RB1 en pacientes colombianos con retinoblastoma esporádico mediante PCR-SSCP seguido de secuenciación.
Materiales y métodos. Se analizaron cuatro pacientes con retinoblastoma esporádico para la detección de variaciones en el gen RB1 mediante PCR-SSCP, seguida de secuenciación.
Resultados. Se identificaron cinco variaciones del gen RB1 : tres mutaciones nuevas (una de línea germinal y dos somáticas), un polimorfismo nuevo y una mutación somática ya reportada. Las cuatro mutaciones se encontraron en tres pacientes con retinoblastoma unilateral y uno con bilateral. La mutación germinal se detectó en un paciente con compromiso unilateral y no se encontró en los padres ni en los tres hermanos analizados.
Conclusión. Estos resultados enfatizan la importancia, para asesoría genética y manejo clínico, de identificar mutaciones del gen RB1 en pacientes con retinoblastoma esporádico. La descripción de una nueva variante en RB1 es interesante, dado el muy bajo número de polimorfismos reportados para este gen.
Palabras clave: retinoblastoma/genética, genes de retinoblastoma, mutación, polimorfismo genético, Colombia.
doi: http://dx.doi.org/10.7705/biomedica.v33i1.643
Retinoblastoma (RB) is an eye cancer of the developing retina in early childhood which has an estimated incidence of 1:20,000 live births. It originates in altered or null retinoblastoma protein (pRb) expression. pRb is a nuclear phosphoprotein of 928 amino acid with an MW of 110 kDa which has a very important role in regulation of the cell cycle G1/S check point (1). pRb is coded by the retinoblastoma gene ( RB1 ), located on chromosome 13q14.2, which was the first tumor suppressor gene identified (2-4). The RB1 gene is 183 kb in length, is composed of 27 exons, and encodes a 4.7 kb mRNA (4). Genetic alterations in both RB1 alleles in the retinal cells are required for the development of retinoblastoma (5).
This tumor has two presentations: sporadic (60%) and familial (40%). In the sporadic form, non-hereditary RB1 gene mutations take place in a single retinoblast cell and are therefore only present in tumor DNA (somatic mutations). Sporadic RB is mainly unilateral, lacks family history, and has no risk of transmission. On the other hand, in the familial form of RB the first mutation is inherited as a germ-line mutation. Therefore all tissues are expected to carry one copy of the mutated RB1 gene. This first mutation can be transmitted to the descendants as a susceptibility risk factor. The second inactivating mutation is somatic and is present only in tumor DNA. Most patients with familial RB have bilateral, multifocal, or early onset tumors (5); however, nearly 15% of familial RB cases present as unilateral tumors (6-10).
The development of sensitive and reliable genetic tests to detect RB1 mutations has improved identification of carriers and facilitated accurate genetic counseling. To date, a total of 1165 unique RB1 DNA variants including polymorphisms and mutations have been registered in the Leiden Open Variation Database (LOVD). RB1 mutations show an extensive heterogeneity without hot spots. Different techniques have been used to identify genetic variations in the RB1 gene. The single strand conformation polymorphism (SSCP) technique has been used to screen for mutations before confirmatory sequencing is performed (11-15). This technique is based on the sequence-dependent migration patterns of single-stranded DNA fragments as they migrate through nondenaturing polyacrylamide gel electrophoresis.
The aim of this study was to identify mutations in the RB1 gene in Colombian patients with sporadic retinoblastoma by PCR-SSCP followed by sequence. In this study we report the identification of three new RB1 gene mutations in four patients with sporadic retinoblastoma in Colombia. We also describe a new RB1 gene variant that increases the small number of polymorphisms reported for this gene.
Materials and methods
Patients
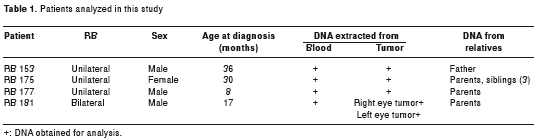
Four sporadic RB patients referred to the Instituto de Genética (Universidad Nacional de Colombia) by the Ophthalmology service of the Instituto Nacional de Cancerología - Bogotá, Colombia, Hospital de La Misericordia and Fundación Carlos Ardila Lulle were analyzed after proper informed consent was obtained and approved by Institutional Review Board (IRB) (Universidad Nacional de Colombia).Three patients had unilateral tumors and one had a bilateral tumor at the time of diagnosis. No previous history of RB was present in any of the patients´ families.
DNA from each patient was obtained from whole blood and from tumors by phenol-chloroform extraction followed by ethanol precipitation. Some formaldehyde fixed and paraffin embedded (FFPE) tumors where extracted following the protocol reported by Goelz et al (16). In addition, DNA was obtained from whole blood from parents and siblings in selected cases (table 1).
DNA amplification
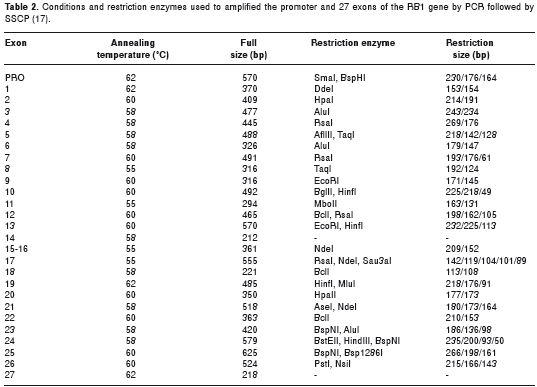
PCR amplification of the promoter and 27 exons of the RB1 gene was performed using primers design by Hogg et al (17) including flanking intron region for each exon that range from 42 to 344pb depending on the exon. Amplification was performed in a 25 µl of total volume containing 100 ng of genomic DNA, 20 nmol of each dNTP, 1 unit of Taq polymerase (Promega Corporation, Madison, WI, USA) and 50 pmol of each primer (Oligo Therapeutics Inc, USA), in standard buffer conditions, except that for the promoter and exon 1 2.5% DMSO was used. Thermal cycling conditions consisting of thirty cycles of denaturing for 30 sec at 94 °C, annealing for 30 sec to 55 °C-62 °C and extension at 72 °C for 1 min were performed in a PTC-100 (MJ Research, Waltham, USA) followed by an additional 10 min at 72 °C. In table 2 we describe the annealing temperatures and the lengths of PCR products.
Single strand conformational polymorphism (SSCP)
In order to optimize SSCP, the large PCR fragments were digested with restriction enzymes as previously described (17) (Table 2). Non-isotopic SSCP analysis was performed in 6-10% polyacrylamide gel containing 5% (v/v) glycerol, in 0.5X TBE buffer. Briefly, 1 µl of amplified products (with or without digestion) was diluted in 3 µl of denaturing buffer (formamide 95%, 10 mM EDTA, 10mM NaOH, 0.05% xylene cyanol, 0.05% bromophenol blue). Immediately prior to loading, diluted samples were denatured at 95°C for three minutes and chilled in ice to minimize renaturation. Samples were electrophoresed at 200 V for 6 hours at room temperature. DNA was visualized by silver-staining. Briefly, gels were fixed for 20 min in 10% (v/v) glacial acetic acid. After three washes with deionized water for 2 min each, gels were stained with 0.2% (w/v) AgNO 3 solution for 20 min, washed with deionized water, and developed in sodium carbonate (30 g/L), 0.4% (v/v) formaldehyde for 10 min.
Sequencing
DNA amplicons that showed bandshift variation in SSCP gels were cloned into the pGEM-T vector and used to transform JM109 competent cells (Promega Corporation, Madison, WI, USA) following manufacturers recommendations. Plasmid DNA isolation was carried out using Wizard Minipreps kits (Promega Corporation, Madison, WI, USA). DNA sequence was carried out using Sequenase T7 version 2.0 kit (Amersham Corporation, Piscataway, NJ 08854 USA), following manufacturers recommendations, and electrophoresed in 6% polyacrylamide gel. Mutations identified were on the RB1 genomic sequence (GenBank accession number: L11910).
Results
An exon by exon SSCP analysis of the RB1 gene followed by sequencing was performed for the four sporadic RB patients. Five RB1 gene variations were found in these four patients (table 1, figure 1). Three of the variations detected were somatic mutations and two were germinal mutations which were detected in both whole blood and tumors. When a germline mutation was identified in a patient, parents and siblings were also analyzed for the same mutation. Four of the five genetic variations detected were new and have not yet been registered in the Leiden Open Variation Database (LOVD) (table 3).

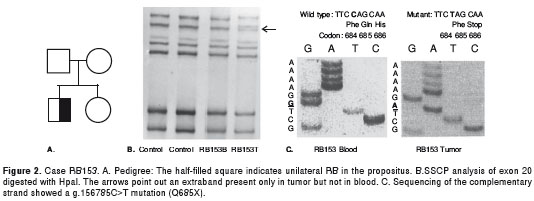
RB 153: The propositus was a boy with unilateral RB diagnosed at three years of age (figure 2A). SSCP of exon 20 showed bandshift in the tumor DNA but not in blood DNA (somatic mutation) (figure 2B). Sequencing showed a g.156785C>T mutation causing a nonsense mutation in codon 685(Q685X) (figure 2C). This mutation had been previously reported by Najera et al (18) in a patient with bilateral RB as a germline mutation.
RB 175: The propositus was a girl with unilateral RB diagnosed at two years of age (figure 3A). SSCP of exon 20 showed alteration in both the tumor and blood (figure 3B y figure 3C). DNA sequencing showed two heterozygous variations in the RB1 gene (figure 3D y 3E). One variation was a C deletion in codon 661, g.156712delC, causing a frameshift mutation introducing a premature stop codon at 662 (figure 3D). This mutation was present in both the patient´s tumor and blood (germline mutation) but absent in her parents and siblings. The second mutation found g.IVS20+2T>G was present only in tumor DNA (somatic mutation). It was a transition in the invariant nucleotide of the splice donor site of intron 20 that probably destroyed this splice donor site (figure 3E).
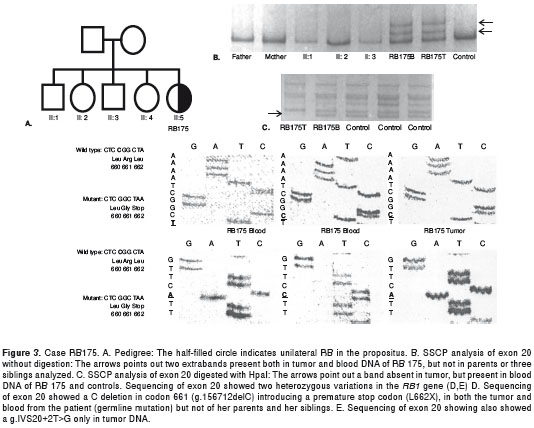
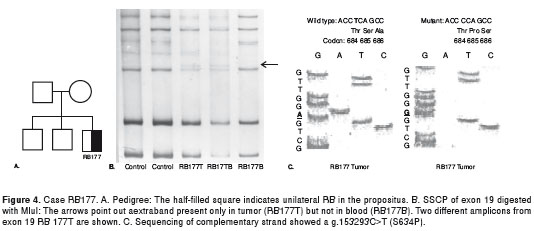
RB 177: The propositus was a boy with unilateral RB diagnosed at eight months of age (figure 4A). SSCP analysis of exon 19 showed motility alteration in tumor DNA (figure 4B). Sequencing showed a g.153293T>C mutation, causing a missense mutation in codon 634 (S634P) (figure 4C). This mutation was heterozygous in the tumor and was not detected in blood (somatic mutation).
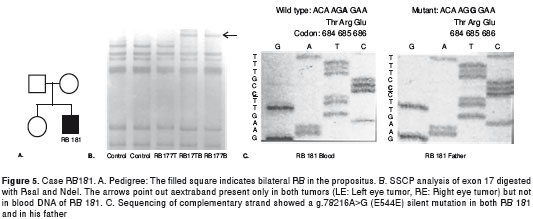
RB 181: The propositus was a boy with bilateral RB diagnosed at seventeen months of age (figure 5A). SSCP analysis of exon 17 showed motility alteration both in blood and tumor DNA (figure 5B). Sequencing showed a silent homozygous g.78216A>G mutation in codon 544 (E544E) (figure 5C). This mutation was also present as a homozygous change in the boy´s father.
Discussion
One of the most important challenges for clinical management of RB patients and their families is to identify genetically predisposed individuals by detection of germline causative mutation in RB patients. Screening for constitutional RB1 mutation should become an integral part of current management of any patient and relatives affected by retinoblastoma. The presence or absence of a germline mutation in RB1 in a patient without family history is very important because distinguish between hereditary and non-hereditary RB and it will supply critical information for prognosis, treatment planning, follow-up care, genetic counseling, presymptomatic diagnosis and enable in uterus screening. Children with heritable RB, in addition to experiencing bilateral vision impairment in early childhood, are at increased risk for developing primitive neuroectodermal tumors and second tumors later in life. The genetically predisposed individuals can be screened ophthalmologically and the tumors treated as they arise.Unequivocal identification of mutant gene carriers will also eliminate the need for costly and time-consuming procedures for family members who are not carriers (19,20). On the other hand, the large size of the RB1 gene (180 kb), its high degree of mutational heterogeneity, the high proportion of mutations arise from small (< 10 bp) sequence changes and the presence of mosaicism and the possibility of mutation within non-coding regions make it very difficult to achieve efficient detection of RB1 mutations for all patients. For these reasons despite the RB1 gene was the first tumor suppressor gene to be identified, many patients and their families have never benefited from genetic testing (21).
In the present study, non-isotopic SSCP and sequencing were used to analyze four sporadic RB patients. Five RB1 gene variants were found in three patients with unilateral RB and one patient with bilateral RB. Four out of five RB1 gene variations detected corresponded to mutations with expected changes in the pRb and the other is a silent mutation that not change de amino acid sequence in the pRb.
Interestingly, of the four mutations detected in our cases, three were novel mutations: g.156712delC (L662X), a frameshift mutation, g.153293T>C (S634P), a missense mutation, and g.IVS20+2T>G, a consensus splice junction sequence nucleotide change (table 3). This emphasizes for the high degree of mutational heterogeneity of this large gene. The additional mutation detected, was a g.156785C>T (Q685X) nonsense somatic mutation, that had been already reported by Najera et al (18). These mutations generate premature stop codons or affect the splicing site of the gene (table 1).
In two patients with unilateral RB, we detect only somatic mutations. The absence of germline mutation in these patients indicates that they correspond to non-hereditary RB and they don´t have risk to transmission for his offspring.
The detection of a germline mutation in unilateral RB175 emphasizes the importance of identifying these mutations for genetic counseling in families with an RB child. This case was a unilateral RB case without family history. However, the mutation detected was a germline mutation not present in any of the relatives. The absence of this mutation in parents and siblings indicates that this mutation may have arisen in the patient herself in the zygote or very early post-zygote stage, or it could represent a germline mosaicism inherited from one of her parents. Recently, Bunin et al (22) reported an association between gonadal radiation exposure in men and women and new germline RB1 mutations detectable in their children. However, no radiation exposure history could be documented in our case. The detection of a germline mutation in this patient with unilateral RB is very important for her management because she has an increased risk to develop a RB in the other eye and is probably that she can develop a second tumor in her life. Since the germline mutation was only detected in the patient, she will have a 50% risk for transmission to her offspring and with de germline causative mutation identified in this study could be possible identified genetically predisposed individuals in her descendants. In RB 175 we also identify a somatic mutation that correspond to second-hit because is only present in tumor.
The other variant was a g.78216A>G in codon 544 (E544E), homozygous silent point mutation present in the RB181 patient and his father which is probably a novel polymorphism. Because it doesn´t change the amino acid codified by codon 544 and it is present in unaffected parent, and doesn´t have a clinical significance. Although the RB1 gene spans over 180 kb, only a few polymorphisms within this locus have been identified. Ten RB1 gene variants without noticeable phenotypic effects are registered in the Leiden Open Variation Database (LOVD). This new polymorphism detected in a Colombian family could represent a variation of the RB1 gene at the population level. In this patient we cannot identify a causative mutation in tumor not in blood. Probably this obeys to sensitivity of SSCP method or the possibility or other kind of alteration that is not possible to detect by methodology used for this study (e.g. large deletion, mutation in non-coding regions situated outside the explored sequences, hypermethylation or other epigenetic alteration) (23). This is a bilateral RB and for this reason his offspring has a 50% to develop RB but in this case is necessary for appropriate genetic counseling perform other techniques that can identify the germline mutation.
There are other studies in Colombia (24) and other Latino American countries as Cuba (24), Argentina (25,26), Brazil (27) and Mexico (14,28) that report, in some cases with almost the same methodology, different mutations, that confirms the high degree of variability of mutational spectrum in RB1 gene.
Many techniques, different of PCR-SSCP, had been use alone or in combination to detected RB1 mutation. These techniques includes: denaturing gradient gel electrophoresis (DGGE)or heteroduplex analysis (12,26), multiplex ligation dependent probe amplification (MLPA) (29) high performance liquid chromatography (HPLC) (6,7), Southern blot (9), fluorescent in situ hybridization (FISH) (9), protein truncation testing (PTT) (8) and PCR-RFLP (10). No single genetic testing method currently available can detect all RB1 mutation types. The most effective and efficient strategies to date have used a series of nested tests. The European Genetics Quality Network has a guideline for molecular analysis of retinoblastoma (13).These protocols, some of which have used as many as 5 different testing modalities, are costly, time consuming and the final sensitivities ranges up to 83% (never 100%) (30). These characteristics reduces clinical utility and is important in the future develop a more efficient method for routine genetic testing with clinical use.
When mutational screening failed to identify the disease-causing mutation the use of polymorphic short tandems (STR) as recommended by the Best Practice Guidelines of the European Molecular Genetics Quality Network (EMQN) for indirect testing of RB1 and useful for establish loss of heterozygosity (LOH). Recently, Munier el at report an algorithm based on linkage analysis that identified patients with true risk (31). High-throughput methodologies as whole-genome sequencing, whole gene sequencing and SNP array methodologies have been used to identify underlying alternative mechanisms of RB genesis not known at present (32).
In conclusion, the present study describes three new RB1 gene mutations with clinical relevance for genetic counseling and clinical management of RB patients, and describes a new RB1 gene variant that increases the small number of polymorphisms detected in this gene.
We would like to thank the Instituto Nacional de Cancerología, Fundación Carlos Ardila Lulle and Hospital de la Misericordia for their assistance in collecting the samples.
None declared.
Instituto de Genética, Universidad Nacional de Colombia, Bogotá D.C., Colombia
Corresponding author: Martha Lucía Serrano, Departamento de Química, Facultad de Ciencias, Universidad Nacional de Colombia, Carrera 30 N° 45-03, edificio 451, oficina 326, Bogotá, D.C., Colombia Telephone: (571) 316 5000, extensión 14435 mlserranol@unal.edu.co
1. Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323-30. http://dx.doi.org/10.1016/0092 -8674(95)90385-2 [ Links ]
2. Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM , et al .A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643-6. http://dx.doi.org/10.1038/323643a0 [ Links ]
3. McGee TL, Yandell DW, Dryja TP.Structure and partial genomic sequence of the human retinoblastoma susceptibility gene. Gene. 1989;80:119-28. http://dx.doi.org/10.1016/0378-1119(89)90256-4 [ Links ]
4. Toguchida J, McGee TL, Paterson JC, Eagle JR, Tucker S, Yandell DW , et al .Complete genomic sequence of the human retinoblastoma susceptibility gene. Genomics. 1993;17:535-43. http://dx.doi.org/10.1006/geno.1993.1368 [ Links ]
5. Knudson AG Jr.Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820-3. [ Links ]
6. Boubekeur A, Louhibi L, Mahmoudi K, Boudjema A, Mehtar N. Etude moleculaire du retinoblastome dans la population algerienne. Recherche de mutations du gene Rb au niveau constitutionnel et tumoral. Bull Cancer. 2012;99:127-35. [ Links ]
7. Houdayer C, Gauthier-Villars M, Lauge A, Pages-Berhouet S, Dehainault C, Caux-Moncoutier V , et al. Comprehensive screening for constitutional RB1 mutations by DHPLC and QMPSF. Hum Mutat. 2004;23:193-202. http://dx.doi.org/10.1002/humu.10303 [ Links ]
8. Tsai T, Fulton L, Smith BJ, Mueller RL, González GA, Uusitalo MS , et al. Rapid identification of germline mutations in retinoblastoma by protein truncation testing. Arch Ophthalmol. 2004;122:239-48. http://dx.doi.org/10.1001/archopht.122.2.239 [ Links ]
9. Brichard B, Heusterspreute M, De Potter P, Chantrain C, Vermylen C, Sibille C , et al. Unilateral retinoblastoma, lack of familial history and older age does not exclude germline RB1 gene mutation. Eur J Cancer. 2006;42:65-72. http://dx.doi.org/10.1016/j.ejca.2005.07.027 [ Links ]
10. Parsam VL, Kannabiran C, Honavar S, Vemuganti GK, Ali MJ. A comprehensive, sensitive and economical approach for the detection of mutations in the RB1 gene in retinoblastoma. J Genet. 2009;88:517-27. [ Links ]
11. Orita M, Suzuki Y, Sekiya T, Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989;5:874-9. http://dx.doi.org/10.1016/0888-7543(89)90129-8 [ Links ]
12. Blanquet V, Turleau C, Gross-Morand MS, Senamaud-Beaufort C, Doz F, Besmond C. Spectrum of germline mutations in the RB1 gene: A study of 232 patients with hereditary and non hereditary retinoblastoma. Hum Mol Genet. 1995;4:383-8. http://dx.doi.org/10.1093/hmg/4.3.383 [ Links ]
13. Lohmann DR, Brandt B, Hopping W, Passarge E, Horsthemke B.The spectrum of RB1 germ-line mutations in hereditary retinoblastoma. Am J Hum Genet. 1996;58:940-9. [ Links ]
14. Macias M, Dean M, Atkinson A, Jiménez-Morales S, García-Vázquez FJ, Saldana-Álvarez Y , et al. Spectrum of RB1 gene mutations and loss of heterozygosity in Mexican patients with retinoblastoma: Identification of six novel mutations. Cancer Biomark. 2008;4:93-9. [ Links ]
15. Sampieri K, Hadjistilianou T, Mari F, Speciale C, Mencarelli MA, Cetta F , et al. Mutational screening of the RB1 gene in Italian patients with retinoblastoma reveals 11 novel mutations. J Hum Genet. 2006;51:209-16. http://dx.doi.org/10.1007/s10038-005-0348-3 [ Links ]
16. Goelz SE, Hamilton SR, Vogelstein B.Purification of DNA from formaldehyde fixed and paraffin embedded human tissue. Biochem Biophys Res Commun. 1985;130:118-26. http://dx.doi.org/10.1016/0006-291X(85)90390-0 [ Links ]
17. Hogg A, Onadim Z, Baird PN, Cowell JK. Detection of heterozygous mutations in the RB1 gene in retinoblastoma patients using single-strand conformation polymorphism analysis and polymerase chain reaction sequencing. Oncogene. 1992;7:1445-51. [ Links ]
18. Najera C, Sánchez F, Mateu E, Prieto F, Beneyto M.Diagnóstico temprano del retinoblastoma: importancia de la búsqueda de mutaciones en el gen RB1. Med Clin (Barc). 2001;116:365-72. [ Links ]
19. Dhar SU, Chintagumpala M, Noll C, Chevez-Barrios P, Paysse EA, Plon SE.Outcomes of integrating genetics in management of patients with retinoblastoma. Arch Ophthalmol. 2011;129:1428-34. http://dx.doi.org/10.1001/archophthalmol.2011.292 [ Links ]
20. Pradhan MA, Ng Y, Strickland A, George PM, Raizis A, Warrington J , et al. Role of genetic testing in retinoblastoma management at a tertiary referral centre. Clin Experiment Ophthalmol. 2010;38:231-6. http://dx.doi.org/10.1111/j.1442-9071.2010.02239.x [ Links ]
21. Tran HV, Schorderet DF, Gaillard MC, Balmer A, Munier FL.Risk assessment of recurrence in sporadic retinoblastoma using a molecular-based algorithm. Ophthalmic Genet. 2012;33:6-11. http://dx.doi.org/10.3109/13816810.2011.610859 [ Links ]
22. Bunin GR, Felice MA, Davidson W, Friedman DL, Shields CL, Maidment A , et al. Medical radiation exposure and risk of retinoblastoma resulting from new germline RB1 mutation. Int J Cancer. 2011;128:2393-404. http://dx.doi.org/10.1002/ijc.25565 [ Links ]
23. Abidi O, Knari S, Sefri H, Charif M, Senechal A, Hamel C , et al. Mutational analysis of the RB1 gene in Moroccan patients with retinoblastoma. Mol Vis. 2011;17:3541-7. [ Links ]
24. Alonso J, Frayle H, Menéndez I, López A, García-Miguel P, Abelairas J , et al. Identification of 26 new constitutional RB1 gene mutations in Spanish, Colombian, and Cuban retinoblastoma patients. Hum Mutat. 2005;25:1-8. http://dx.doi.org/10.1002/humu.9299 [ Links ]
25. Parma DL, Dalamon VK, Fernández C, Szijan I, Damel A.Importancia de los estudios de biología molecular en el asesoramiento genético de familias argentinas con retinoblastoma. Arch Soc Esp Oftalmol. 2009;84:557-62. http://dx.doi.org/10.4321/S0365-66912009001100004 [ Links ]
26. Fernández C, Repetto K, Dalamon V, Bergonzi F, Ferreiro V, Szijan I.RB1 germ-line deletions in Argentine retinoblastoma patients. Mol Diagn Ther. 2007;11:55-61. [ Links ]
27. De Andrade AF, Da Hora Barbosa R, Vargas FR, Ferman S, Eisenberg AL, Fernandes L , et al. A molecular study of first and second RB1 mutational hits in retinoblastoma patients. Cancer Genet Cytogenet. 2006;167:43-6. http://dx.doi.org/10.1016/j.cancergencyto.2005.08.017 [ Links ]
28. Rodríguez M, Salcedo M, González M, Coral-Vázquez R, Salamanca F, Arenas D. Identification of novel mutations in the RB1 gene in Mexican patients with retinoblastoma. Cancer Genet Cytogenet. 2002;138:27-31. http://dx.doi.org/10.1016/S0165-4608(02)00577-0 [ Links ]
29. Sellner LN, Edkins E, Smith N. Screening for RB1 mutations in tumor tissue using denaturing high performance liquid chromatography, multiplex ligation-dependent probe amplification, and loss of heterozygosity analysis. Pediatr Dev Pathol. 2006;9:31-7. http://dx.doi.org/10.2350/07-05-0078.1 [ Links ]
30. Nichols KE, Houseknecht MD, Godmilow L, Bunin G, Shields C, Meadows A , et al. Sensitive multistep clinical molecular screening of 180 unrelated individuals with retinoblastoma detects 36 novel mutations in the RB1 gene. Hum Mutat. 2005;25:566-74. http://dx.doi.org/10.1002/humu.20184 [ Links ]
31. Lohmann D, Scheffer H, Gallie B. Best practice guideline for molecular analysis of retinoblastoma. European Molecular Genetics Quality Network. Fecha de consulta: 24 de julio del 2012. Disponible en: http://www.emqn.org/emqn/digitalAssets/0/239_RB.pdf . [ Links ]
32. Zhang J, Benavente CA, McEvoy J, Flores-Otero J, Ding L, Chen X , et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329-34. http://dx.doi.org/10.1038/nature10733 [ Links ]













