Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Biomédica
versão impressa ISSN 0120-4157
Biomédica vol.33 no.4 Bogotá out./dez. 2013
https://doi.org/10.7705/biomedica.v33i4.1678
ARTÍCULO ORIGINAL
doi: http://dx.doi.org/10.7705/biomedica.v33i4.1678
1 División de Enfermedades Infecciosas, Centro Nacional de Investigaciones Científicas, La Habana, Cuba
2 Centro de Estudios de Proteínas, Facultad de Biología, Universidad de La Habana, La Habana, Cuba
3 Departamento de Gastroenterología, Hospital Carlos J. Finlay, La Habana, Cuba
Author contributions:
L. González and B. L. Rodríguez: contributed equally to this work. Designed the research, analyzed the data and wrote the paper.
K. Marrero: designed the research
L. Martínez, O. Reyes and E. Rodríguez: collected the samples and managed the patient data.
L. González and E. Rodriguez: performed the research.
Recibido: 10/10/12; aceptado:27/05/13
Introduction: Helicobacter pylori strains expressing cytotoxic CagA protein are more commonly associated with peptic ulceration, atrophic gastritis and gastric adenocarcinoma than those lacking CagA. Determination of anti-CagA antibodies, therefore, acquires a relevant clinical significance in the serological detection of H. pylori infection and disease risk prediction. However, the CagA-serology has been questioned due to the differences found in their performance evaluations in different populations.
Objective: To obtain a recombinant CagA fragment useful for serodiagnosis of H. pylori infection
Methods: A fragment of the cagA gene was cloned into a prokaryotic T7 RNA polymerase expression vector. A recombinant C-terminal His 6 -tagged CagA was expressed, subsequently solubilized with urea and purified by immobilized metal affinity chromatography. The performance of the recombinant protein was evaluated using 180 human serum samples with an in-house Western blot assay compared to the Helicoblot 2.1 reference test.
Results: The expressed His 6 -tagged CagA showed an immunoreactive 80kDa band as was revealed by SDS-PAGE and Western blot analysis using two different specific anti-CagA polyclonal antibodies.
The recombinant protein was successfully purified obtaining a 93% of purity. The performance analysis of the purified recombinant antigen showed good immunoreactivity and exhibited values of sensitivity, specificity and accuracy of 88.1%, 100% and 92.7%, respectively.
Conclusion: The CagA fragment of the study may constitute a useful tool for serological diagnosis of CagA-positive H. pylori infection.
Keywords: Helicobacter pylori ; serological test
doi: http://dx.doi.org/10.7705/biomedica.v33i4.1678
Clonación y expresión de un fragmento recombinante del gen cagA de Helicobacter pylori y su evaluación preliminar en el serodiagnóstico
Introducción. Las cepas de Helicobacter pylori que expresan la citotoxina CagA, se asocian más frecuentemente con úlcera péptica, gastritis atrófica y adenocarcinoma gástrico que las que carecen de esta citotoxina. Por lo anterior, el determinar la presencia de anticuerpos anti-CagA adquiere gran importancia clínica en la detección serológica de la infección por H. pylori y la predicción del riesgo de enfermedades. Sin embargo, los métodos serológicos que emplean CagA han sido cuestionados debido a las diferencias encontradas en las evaluaciones de su desempeño en diversas poblaciones.
Objetivo. Obtener un fragmento recombinante de la proteína CagA para el serodiagnóstico de la infección por H. pylori .
Materiales y métodos. Un fragmento del gen cagA fue clonado en un vector de expresión procariota que contenía el promotor de la T7 ARN polimerasa. El fragmento de la proteína CagA con seis histidinas en la región C-terminal, se expresó, se solubilizó con urea y se purificó por cromatografía de afinidad con iones metálicos inmovilizados. El desempeño de la proteína recombinante se evaluó empleando un método in house de Western Blot y 180 sueros humanos. Los resultados se compararon con la prueba de referencia Helicoblot 2.1.
Resultados. La proteína CagA expresada mostró una banda inmunorreactiva de 80 kDa en el Western Blot al emplear dos anticuerpos policlonales anti-CagA específicos. La proteína recombinante fue purificada hasta un 93 % de pureza y el análisis de desempeño del antígeno recombinante purificado mostró buena inmunorreacción y exhibió valores de sensibilidad, especificidad y exactitud de 88,1 %, 100 % y 92,7 %, respectivamente.
Conclusiones. El fragmento de la proteína CagA del estudio puede constituir una herramienta útil para el diagnóstico serológico de la infección por cepas de H. pylori positivas para CagA.
Palabras clave: Helicobacter pylori , pruebas serológicas.
doi: http://dx.doi.org/10.7705/biomedica.v33i4.1678
Introduction
Helicobacter pylori is a Gram-negative spiralshaped bacterium that persistently colonizes the stomach of more than half of the human population. This persistent colonization plays an important role in the appearance of gastroduodenal pathologies, such as peptic ulcers, gastric adenocarcinoma and gastric mucosa-associated lymphoid tissue (MALT) lymphoma (1).
The persistence of this infection, which is crucial for developing chronic disease phenotypes, mainly depends on specific characteristics of the bacterium. The most important virulence factor of H. pylori is the cytotoxin-associated antigen CagA. This protein is coded by the cagA gene that is part of the cag-pathogenicity island which encodes the components of a type IV secretion system (T4SS). H. pylori uses this T4SS to inject CagA into human gastric cells, where it perturbs several host signaling pathways to provide a local environment that is more suitable for the pathogen survival and thereby promoting severe damages of gastric epithelial cells and mucosa (2,3,4).
The CagA protein is an immunodominant antigen (5,6) and the cagA -positive strains are commonly associated with more severe gastroduodenal diseases than cagA -negative strains (7,8). Several studies in cell culture and animal models indicate the likely role of CagA in human cancer associated with H. pylori infection (3). Transgenic expression of CagA in mice led to the development of gastric epithelial hyperplasia and adenocarcinoma of the stomach and the small intestine, providing the first direct evidence of the potential oncogenicity of CagA in vivo (9). Thus, CagA is the first bacterial oncoprotein to be identified in relation to human cancer. Therefore, determination of the CagAstatus in H. pylori infection is an important and an informative approach in disease risk prediction.
Different methods have been used to detect H. pylori infection with CagA-positive strains. Among all, the serologic methods have been successfully used due to the immunogenicity of this protein and the non- invasivity of the method (10). In addition, it has been observed that CagA seropositivity is able to predict the development of atrophy and constitute a risk factor for intestinal metaplasia (11). However, the accuracy of CagA-serology has been challenged due to the differences found in their evaluations. It has been suggested that this variability is mainly related with the differences in the antigens or methodologies used to detect the anti-CagA antibodies, or the use of different populations (12). Here we described a new recombinant fragment of CagA and evaluated its efficacy for the serodiagnosis of the CagA status of H . pylori infection.
Materials and methods
Serum samples
Serum samples were obtained from 180 consecutive Cuban dyspeptic patients with an average age of 36.5 years (range, 18 to 55) who attended the Department of Gastroenterology of Carlos J. Finlay Hospital. Ninety-two of the patients presented gastritis (51.1 %), 34 gastric ulcer (18.8 %) and 54 duodenal ulcer (30.1 %). All patients gave their written informed consent to participate in the study and the Carlos J. Finlay Hospital ethic committee approved the protocol by which the specimens were obtained.
Bacterial strains, plasmids and reagents
H. pylori strain 26695 was used for genomic DNA extraction. E. coli Mach1 (Invitrogen, USA) was used as a host strain for cloning manipulation of cagA gene. E. coli BL21(DE3) (Invitrogen, USA) was used as CagA expression strain. The pET22b(+)plasmid (Novagen, USA) was used as cloning and expression vector. This vector contains the T7 promoter and allows expression of the recombinant protein fused to histidine tail at the C-terminus. The restriction enzymes and PCR reagents used in cloning procedures were purchased from Roche, USA.
Molecular techniques and construction of the expression plasmid
Genomic DNA from H. pylori was extracted by CTAB methodology with phenol/chloroform and isopropanol precipitation as previously described (13). A fragment of the cagA gene was amplified by PCR using forward primer cagA.F [5´-CATGCCA TGGGGGATAACAGGCAAGCTTTTGA-3´] which created the NcoI site (nderlined), and reverse primer cagA.R [5´-GAATTCTCGAGGTCGCTTTTTGC-3´] XhoI site These primers were designed based on the DNA sequences of 26695 cagA gene obtained from the GenBank database and using the Gene Runner software version 3.05.
PCR reactions were performed in final volumes of 50 µ l containing 100 ng of H. pylori genomic DNA, 0.2 mM dNTP, 2.5 mM MgCl 2, 0.6 µ M of each primer, 1 unit of Taq DNA polymerase and 1× reaction buffer. For amplification, a Thermal Cycler, Mastercycler® personal (Eppendorf, Germany) was used with the following settings: 1 min at 94 °C followed by 40 cycles of 1 min at 94 ºC, 1 min at 61 °C and 1 min at 72 °C and then a final extension of 5 min at 72 °C. The PCR products were analyzed by agarose (2%) gel electrophoresis using a 100 bp DNA molecular size marker (Roche, USA) and then purified using the High Pure PCR Product Purification Kit (Roche, USA).
The purified PCR product was cloned into the pET22b(+) expression vector following the manufacturer instructions. The cloning product was transformed into E. coli Mach1 electrocompetent cells and recombinant clones were identified by plasmid purification using the High Pure Plasmid Isolation Kit (Roche, USA) and confirmed by restriction enzyme analysis. The identity of the insert was determined by DNA sequencing (Macrogen Inc., Korea). Sequencing reactions were performed for both DNA strands, and the ClustalW2 multiplesequence alignment software was used to analyze sequencing results.
Expression and localization of recombinant CagA
Recombinant plasmid pET22b(+)-CagA was transformed into E. coli BL21(DE3) electrocompetent cells and recombinant clones were identified as described above. One recombinant clone was used to inoculate 5 ml of Luria–Bertani medium containing 100 µ g/mL ampicillin which was incubated in an orbital shaker (37 ºC, 200 rpm) until OD 600nm 0.6. At this moment, expression of the recombinant protein was induced with isopropylb- D-1-thiogalactopyranoside (IPTG), with a final concentration of 0.1 mM, and incubated for additional 4 h in the same conditions. Cells were harvested by centrifugation (10 000 g, 4 °C, 5 min) and resuspended in 0.1 M phosphate-buffered saline, pH 7.4 (PBS) and later the 5× SDS-PAGE loading buffer was added (250 mM Tris–HCl pH 6.8, 50% glycerol, 1 M 2-mercaptoethanol and 1% SDS). After boiling for 10 min, the sample was submitted to 12% SDS-PAGE according to Laemmli (14). Expression of the recombinant protein was visualized by Coomassie Blue staining. To confirm the presence and immunorreactivity of the recombinant protein, Western blot analysis was carried out essentially as Towbin and Gordon (15) and two different anti-CagA polyclonal antibodies, kindly provided by professors Guillermo Pérez- Pérez (New York University, School of Medicine, New York, USA) and Rainer Haas (Ludwig Maximilian University, Munich, Germany), were used.
In order to improve the CagA expression, several experimental parameters were optimized: temperature, IPTG concentration, OD 600nm at induction, and incubation time after induction. This protein expression analysis was monitored by SDSPAGE and using a Gene Genius gel documentation system (Syngene, UK).
To localize the recombinant CagA protein (rCagA) into the cell extract, the clone expressing the rCagA was tested regarding the solubility of the protein. Cells were lysed by freeze-thawed three times in buffer A (20 mM Tris, pH 7, containing 0.5 M NaCl, 5 mM imidazole, and 1 mM PMSF) at a ratio of 1 g of wet cells/5 ml of buffer A, as previously described (16). The soluble and insoluble fractions were separated by centrifugation at 10 000 g for 5 min at 4 °C and applied in a 12% SDS–PAGE as above.
Purification of recombinant CagA
The cell lysis insoluble fraction, containing the rCagA, was suspended in buffer A and treated with different concentrations of Urea (2, 4, 6, and 8 M) employing an Ultra-Turrax homogenizer (IKA-WERKE, Germany) at 9500 rpm for 1min. 4 °C, the samples were centrifuged at 14 000 g for 20 min at 4 °C and all soluble and insoluble fractions were analyzed by 12% SDS–PAGE to determine the minimal Urea concentration needed for the best rCagA solubilization. Then, the recombinant protein was purified by immobilized metal affinity chromatography (IMAC) using a Quelating Sepharose Fast Flow matrix (Amersham Pharmacia, Sweden) pre-charged with Cu 2+ . The column was equilibrated with buffer A containing 4 M Urea and the protein refolding was performed oncolumn using a linear gradient from 4 to 0 M Urea in buffer A. The recombinant protein was eluted with a linear gradient from 5 to 200 mM imidazole and fractions of 1 ml were collected and analyzed by 12% SDS–PAGE. The purified protein was dialyzed on Sephadex G-25 column equilibrated with PBS.
Determination of protein concentration and purity
The concentration and purity of the rCagA was determined by Bradford´s assay (17) and SDSPAGE analysis using the Syngene Gene Genius imaging system, UK, respectively. Bovine serum albumin was used as standards in both cases.
Evaluation of recombinant CagA antigen for serodiagnosis
In order to assess the efficacy of the rCagA in serodiagnostic, a panel of 180 human sera was simultaneously evaluated using an in-house Western blot assay and the commercial immunoblot assay Helicoblot 2.1 (HB 2.1) (Genelabs Diagnostics, Singapore), which was regarded as the reference assay in the study. The Western-rCagA was performed as follow: the proteins resolved on a 12% SDS-PAGE gel were transferred to nitrocellulose membrane (Amersham Pharmacia, Sweden) for 3 h at 36V using a wet tank transfer system (Hoefer Inc., USA). The membranes were incubated with blocking reagent (5% skim milk in PBS), 1h at 4°C and subsequently incubated with the serum samples overnight in gentle agitation at room temperature. All sera were diluted 1/100 in PBS containing 0.05% Tween 20. After washing with distilled water containing 0.05% Tween 20, membranes were incubated 1 h at room temperature with a specific anti-human immunoglobulin antibody conjugated to horseradish peroxidase (Promega, USA) diluted 1/5000 in PBS containing 1% skim milk and 0.05% Tween 20. Reacting bands were detected using 3,3´-diaminobenzidine substrate 0.02% in buffer 10mM Tris, pH 7.6.
The HB 2.1 assay was developed in accordance with the manufacturer´s instructions. Results from Western-rCagA test were compared with those from the HB 2.1 test to assess the performance of the recombinant antigen by determining the relative sensitivity, specificity and accuracy as described below,
Sensitivity = a / (a + c) x 100, where ´a´ is the number of sera positive by both, the Western-rCagA and HB 2.1 while ´c´ is the number of sera positive by HB 2.1 but negative by Western-rCagA.
Specificity = d / (b + d) x 100 where ´d´ is the number of sera negative by both, the Western-rCagA and HB 2.1 while ´d´ is the number of sera negative by HB 2.1 but positive by Western-rCagA.
Accuracy = (a + d) / (a + b + c + d) x 100.
Results
Cloning of H. pylori CagA gene
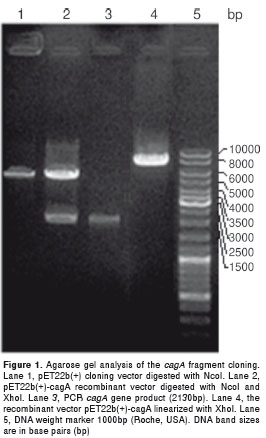
The designed primers allowed effective amplification of the selected fragment of cagA gene. These PCR reactions generated DNA fragments of 2130 bp as it was expected. Amplified fragment digested with NcoI and XhoI, was purified and successfully cloned into the pET22b(+) expression vector as was corroborated by restriction enzyme analysis (Fig. 1). Sequencing of the pET22b(+) insert, compared to 26695 cagA gene, obtained from the GenBank database, confirmed the identity of the cloned cagA sequence (data not shown).
Expression of recombinant CagA
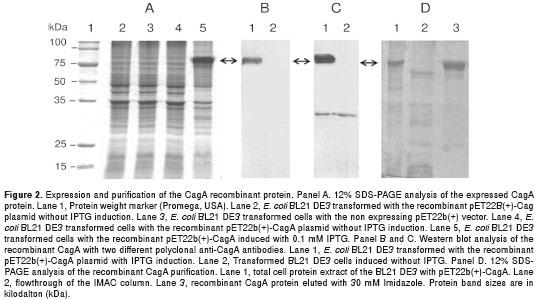
E. coli BL21(DE3) harbouring plasmid pET22b(+)- CagA, expressed a major protein of approximately 80kDa upon IPTG induction that could be detected by SDS–PAGE analysis (Fig. 2A, lane 5). This molecular size corresponds to the estimated molecular weight of the selected cagA fragment (81.5kDa). After SDS-PAGE analysis, the target protein was further analyzed by immunoblotting with two different anti-CagA polyclonal antibodies to corroborate the presence of the recombinant protein. The Western blot assay also showed a major reacting band of 80kDa approximately (Fig. 2B, lane 1). This band was not detected without IPTG induction (Fig. 2B, lane 2).
The optimization of the recombinant protein production showed that the best conditions were an expression at a OD 600nm 0.8 with 1mM IPTG at 37°C and 200 rpm during 4 h. The expressed protein under these conditions constituted 27% of the total cellular protein as was determined by densitometry. The cells were successfully broken by the freeze-thaw procedure and the rCagA was found in the insoluble fraction after cell lysis (data not shown).
Purification of recombinant CagA
The recombinant protein accumulated in the insoluble fraction of cell lysate was solubilized in buffer containing urea. The solubilization assays allowed determining that 4 M urea was the minimal concentration needed to suspend the rCagA with the minimum presence of contaminants (data not shown).
The recombinant protein was successfully purified by IMAC and the rCagA was eluted from the column at 30 mM of imidazole with approximately 93% purity (Fig. 2D, lane 3).
Performance of the Western-rCagA
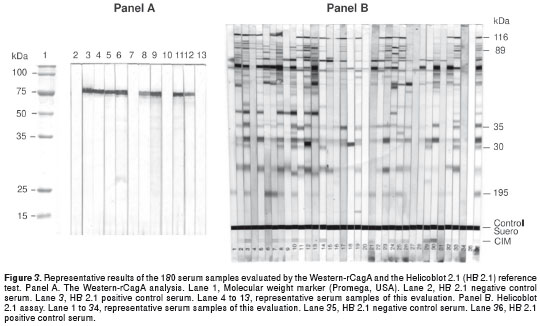
The performance of the rCagA in serodiagnostic was studied using a panel of 180 serum samples and the HB 2.1 as the reference test. Among all sera tested, the HB 2.1 identified 109 (60.6%) CagA positive and 71 (39.4%) CagA negative cases. On the other hand, 96 from the 109 CagA positive sera detected by HB 2.1 were also immunorreactive with the rCagA, while all the 71 CagA negative cases detected by HB 2.1 were also negative by the Western-rCagA. Thus, there were 13 false negative and none false positive results when the Western-rCagA was used to detect anti-CagA antibodies. Therefore, the Western blot assay using the rCagA developed here showed a sensitivity, specificity, and accuracy of 88.1%, 100%, and 92.7%, respectively.
Discussion
Several studies using diagnostic methods to detect CagA antibodies have demonstrated that this antigen improve the sensitivity of H. pylori detection (18). Also, it has been observed that the positivity for CagA antibodies has the ability to predict the development of atrophy and appears related with young gastric cancer cases (10). However, the accuracy of the tests developed with this regard has been questioned because the results differed when they were used in different populations. It has been suggested that an important issue to this heterogeneity appeared to result from differences in the antigens used to detect anti-CagA antibodies (12). Consequently, new studies using the already existent CagA antigens, and/or some new design fragments of the cytotoxin, should be done to evaluate the reactivity of representative populations to different antigens.
In the present study a new recombinant fragment of CagA cytotoxin was expressed in E. coli . The goal of the study has been the production of a specific antigen that should be able to react with a broad range of human anti-CagA antibodies and could improve the serodiagnosis of the CagA-positive H. pylori infection.
The selected fragment corresponds to all conserved N-terminal and central conserved regions of the CagA protein, containing 716 aa and an estimated molecular size of 81.5 kDa. Previous studies have demonstrated that CagA is a challenging protein for recombinant expression, due to a propensity for degradation (6,19). Specifically, it has been suggested that the region preceding the first EPIYA motif contains an important cleavage site (19). We designed a region that excluded both, this site and the CagA variable region at the C-terminus (the EPIYA region) and also lacked the most proximal N- terminal variable region. The selected CagA sequence constitutes a suitable fragment for the recognition of antibodies due to contain several epitopes that are highly conserved and immunogenic (20,21). In agreement with the above data, the selected recombinant CagA fragment of this study showed a low degree of degradation and was highly immunoreactive (Fig. 2 and Fig. 3).
For large-scale production of recombinant proteins, the T7-Lac promoter based vectors containing histidine tails have been successfully used for hyper-expression of the selected proteins, which help in devising simple chromatography protocols for obtaining highly pure protein in high yields (22). The expression system used in this study produced a large amount of the heterologous protein (27% of the total cellular protein). The rCagA was expressed as C-terminally His 6 -tagged protein and was purified by IMAC. The use of the histidine affinity tag facilitated the recovery of the recombinant protein, using one-step chromatography procedure, to obtain a 93% of purity (Fig. 2, panel D) without affecting its immunoreactive properties (Fig. 3, panel A).
The two polyclonal anti-CagA antibodies used here were generated using two different recombinant CagA fragments (20,23). These two recombinant proteins comprise different fragments, which are mostly included in the wide conserved sequence selected for our rCagA. Therefore, the good reactivity observed with both antibodies demonstrated the identity of the CagA produced fragment (Fig. 2, panel B and C).
The CagA seropositivity (60.6%) obtained with Helicoblot 2.1 test in this study was similar to the positivity observed in CagA genotyping of Western strains (24) and slightly lower than the cagA positive strains detected in Cuban H. pylori isolates (25). The Western blot assay with the rCagA developed here demonstrated adequate performance in comparison to HB 2.1 that contains the entire CagA antigen. This comparison showed sensitivity of 88.1%, specificity of 100% and accuracy of 92.7%. The sensitivity was the lowest performance parameter observed, but it is in the accepted range compared with others ELISA and immunoblotting tests developed to detect CagA antibodies (26,27). Interesting, among the 13 false negative samples obtained with the Western-rCagA, 9 were classified as H. pylori negative CagA positive by HB 2.1. It is though that CagA seropositivity in H. pylori -seronegative subjects may either be a sign of a past H. pylori infection due to the long life of anti-CagA antibodies (18,28) or represent a false positive reaction (29). Therefore, probably the rCagA of this study may not contain the epitopes causing either of the above mentioned events that should be, if demonstrated, an advantage of this recombinant fragment to diagnose CagA-positive H. pylori infection. In addition, our rCagA should be sensitive enough due to the fact that contains the conserved part of the CagA cytotoxin, which has been recently shown to have the most reactive epitopes of the entire CagA protein (21). On the other hand, there were no false positive results and therefore the Western-rCagA showed 100% specificity. This is also a good result for the rCagA of this study because the lack of specificity of anti- CagA tests has been attributed in part to differences in the CagA antigens used (12,30).
Taken together, the data of this study suggest that the recombinant CagA fragment developed here should be a good reagent to be included in a CagA serodiagnostic assay to be standardized and validated in representative populations.
The authors have declared no conflicts of interest.
This work was supported by a grant from the National Center for Scientific Research, Havana, Cuba.
Corresponding author
Boris Luis Rodríguez, Centro de Estudios de Proteínas, Facultad de Biología, Universidad de La Habana, Calle 25, Nº 455, Vedado, Plaza de la Revolución, La Habana, Cuba, 10400 Teléfono: (537) 832 4830 rodriguez@fbio.uh.cu
1. Gisbert JP. Helicobacter pylori -related diseases: Dyspepsia, ulcers and gastric cancer. Gastroenterol Hepatol. 2011;34(Suppl.2):15-26. http://dx.doi.org/10.1016/S0210-5705(11)70017-6. [ Links ]
2. Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asaka M, et al. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science.2002;295:683-6. http://dx.doi.org/10.1126/science.1067147 [ Links ]
3. Hatakeyama M. Linking epithelial polarity and carcinogenesis by multitasking Helicobacter pylori virulence factor CagA. Oncogene. 2008;27:7047-54. http://dx.doi.org/10.1038/onc.2008.353. [ Links ]
4. Hatakeyama M. Anthropological and clinical implications for the structural diversity of the Helicobacter pylori CagA oncoprotein. Cancer Sci. 2011;102:36-43. http://dx.doi.org/10.1111/j.1349-7006.2010.01743.x. [ Links ]
5. Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA . 1993;90:5791-5. [ Links ]
6. Tummuru MK, Cover TL, Blaser MJ. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori : Evidence of linkage to cytotoxin production. Infect Immun. 1993;61:1799-809. [ Links ]
7. Nomura AM, Pérez-Pérez GI, Lee J, Stemmermann G, Blaser MJ. Relation between Helicobacter pylori cagA status and risk of peptic ulcer disease. Am J Epidemiol. 2002;155:1054-9. http://dx.doi.org/10.1093/aje/155.11.1054 [ Links ]
8. Camorlinga-Ponce M, Flores-Luna L, Lazcano-Ponce E, Herrero R, Bernal-Sahagún F, Abdo-Francis JM, et al. Age and severity of mucosal lesions influence the performance of serologic markers in Helicobacter pylori -associated gastroduodenal pathologies. Cancer Epidemiol Biomarkers Prev.2008;17:2498-504. http://dx.doi.org/10.1158/1055-9965.EPI-08-0289 [ Links ]
9. Ohnishi N, Yuasa H, Tanaka S, Sawa H, Miura M, Matsui A, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci USA. 2008;105:1003-8. http://dx.doi.org/10.1073/pnas.0711183105 [ Links ]
10. Monteiro L, Oleastro M, Lehours P, Mégraud F. Diagnosis of Helicobacter pylori infection. Helicobacter. 2009;14(Suppl.1):8-14. http://dx.doi.org/10.1111/j.1523-5378.2009.00707.x. [ Links ]
11. Vorobjova T, Maaroos HI, Uibo R. Immune response to Helicobacter pylori and its association with the dynamics of chronic gastritis in the antrum and corpus. APMIS. 2008;116:465-76. http://dx.doi.org/10.1111/j.1600-0463.2008.00934.x [ Links ]
12. Shiota S, Matsunari O, Watada M, Yamaoka Y. Serum Helicobacter pylori CagA antibody as a biomarker for gastric cancer in east-Asian countries. Future Microbiol. 2010;5:1885-93. http://dx.doi.org/10.2217/fmb.10.135. [ Links ]
13. Ausubel F, Brent MR, Kingston RE, Moore DD, Seidman JG, Smith JA et al. Short protocols in molecular biology. New York: John Wiley & Sons Inc; 1995. [ Links ]
14. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680-5. http://dx.doi.org/10.1038/227680a0 [ Links ]
15. Towbin H, Gordon J. Immunoblotting and dot immunobinding –current status and outlook. J Immunol Methods. 1984;72:313-40. http://dx.doi.org/10.1016/0022-1759(84)90001-2 [ Links ]
16. Amersham Biosciences. Purification of (His) 6 -tagged proteins using HiTrap Chelating HP columns charged with different metal ions. Application note, Affinity chromatography. 2002;11;1-8. [ Links ]
17. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248-54. [ Links ]
18. Veijola L, Oksanen A, Sipponen P, Rautelin H. Evaluation of a commercial immunoblot, Helicoblot 2.1, for diagnosis of Helicobacter pylori infection. Clin Vaccine Immunol. 2008;15:1705-10. http://dx.doi.org/10.1128/CVI.00165-08. [ Links ]
19. Angelini A, Tosi T, Mas P, Acajjaoui S, Zanotti G, Terradot L, et al. Expression of Helicobacter pylori CagA domains by library-based constructed screening. FEBS J. 2009;276:816-24. http://dx.doi.org/10.1111/j.1742-4658.2008.06826.x [ Links ]
20. Blaser MJ, Pérez-Pérez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111-5. [ Links ]
21. Klimovich AV, Samoylovich MP, Gryazeva IV, Terekhina LA, Suvorov AN, Klimovich VB. Development of immunoreagents for diagnostics of CagA-Positive Helicobacter pylori infections. Helicobacter. 2010;15:193-200. http://dx.doi.org/10.1111/j.1523-5378.2010.00754.x. [ Links ]
22. Graslünd S, Nordlund P, Weigelt J, Bray J, Gileadi O, Knapp S, et al. Protein production and purification. Nat Methods. 2008;5:135-46. http://dx.doi.org/10.1038/nmeth.f.202. [ Links ]
23. Hohlfeld S, Pattis I, Püls J, Plano GV, Haas R, Fischer W. A C-terminal translocation signal is necessary, but not sufficient for type IV secretion of the Helicobacter pylori CagA protein. Mol Microbiol. 2006;59:1624-37. http://dx.doi.org/10.1111/j.1365-2958.2006.05050.x [ Links ]
24. Murata-Kamiya N. Pathophysiological functions of the CagA oncoprotein during infection by Helicobacter pylori . Microbes Infect. 2011;13:799-807. http://dx.doi.org/10.1016/j.micinf.2011.03.011 [ Links ]
25. Torres LE, Melian K, Moreno A, Alonso J, Sabatier CA, Hernández M, et al. Prevalence of vacA, cagA and babA2 genes in Cuban Helicobacter pylori isolates. World J Gastroenterol. 2009;15:204-10. http://dx.doi.org/10.3748/wjg.15.204 [ Links ]
26. Vaira D, Malfertheiner P, Megraud F, Axon AT, Deltenre M, Hirschl AM, et al. Diagnosis of Helicobacter pylori infection with a new non-invasive antigen-based assay. HpSA European study group. Lancet. 1999;354:30-3. http://dx.doi.org/10.1016/S0140-6736(98)08103-3 [ Links ]
27. Shimoyama T, Fukuda S, Nakasato F, Yoshimura T, Mikami T, Munakata A. Relation of CagA seropositivity to cagPAI phenotype and histological grade of gastritis in patients with Helicobacter pylori infection. World J Gastroenterol. 2005;11:3751-5. [ Links ]
28. Ekström AM, Held M, Hansson LE, Engstrand L, Nyrén O. Helicobacter pylori in gastric cancer established by CagA immunoblot as a marker of past infection. Gastroenterology. 2001;121:784-91. http://dx.doi.org/10.1053/gast.2001.27999 [ Links ]
29. Siman JH, Engstrand L, Berglund G, Floren CH, Forsgren A. Evaluation of western blot CagA seropositivity in Helicobacter pylori seropositive and seronegative subjects. Clin Diagn Lab Immunol. 2005;12:304-9. http://dx.doi.org/10.1128/CDLI.12.2.304-309.2005 [ Links ]
30. Park CY, Cho YK, Kodama T, El-Zimaity HM, Osato MS, Graham DY. New serological assay for detection of putative Helicobacter pylori virulence factors. J Clin Microbiol. 2002;40:4753-6. http://dx.doi.org/10.1128/JCM.40.12.4753-4756.2002 [ Links ]













