Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Biomédica
versión impresa ISSN 0120-4157
Biomédica vol.33 supl.1 Bogotá set. 2013
https://doi.org/10.7705/biomedica.v33i0.731
COMUNICACIÓN BREVE
doi: http://dx.doi.org/10.7705/biomedica.v33i0.731
1 Grupo de Investigaciones en Enfermedades Tropicales, Departamento de Medicina, Universidad del Norte, Barranquilla, Colombia
2 Saskatchewan Disease Control Laboratory, Regina, Saskatchewan, Canada
3 Instituto de Investigaciones Biológicas del Trópico, Universidad de Córdoba, Montería, Colombia
Author contributions:
Andrew K. Falconar designed the study
Margarett Cuello, Virginia Rodríguez, Alfonso Calderón, Germán Arrieta and Salim Mattar performed the Leptospira spp. isolations and the PCRs.
Dorothy Thiry and Paul N. Levett provided training in the PFGE and result analysis and prepared the figures.
Andrew K. Falconar wrote the article.
Claudia M. Romero-Vivas participated in all activities.
Recibido: 03/05/12; aceptado:18/10/12
Introduction: Leptospirosis is a bacterial disease transmitted directly or indirectly from animals to humans that may result in severe hemorrhagic, hepatic/renal and pulmonary disease. There are 20 known Leptospira species and hundreds of serovars, some of which belong to different species. It is essential to identify pathogenic Leptospira serovars and their potential reservoirs to prepare adequate control strategies.
Objective: To characterize the Leptospira serovars isolated from rodents, dogs, pigs and water samples in Colombia.
Materials and methods: Leptospira organisms were isolated and cultured, and pathogenic strains were identified using a polymerase chain-reaction (PCR). Leptospira DNA and Salmonella Braenderup H9812 (molecular weight standard) DNA were cleaved using NotI and subjected to pulsed-field gel electrophoresis (PFGE). The PFGE patterns were analyzed based on bacterial strain-typing criteria and Dice coefficients (DCs) between these isolates and over 200 Leptospira organisms isolated from other parts of the world.
Results: All of the isolates were pathogenic strains, and five were genetically characterized. The P275 (84% DC) and P282 (95% DC) pig isolates were related to the Leptospira interrogans Pomona serovar; the I15 (DC: 100%) rat isolate was identical to the Leptospira interrogans Icterohameorrhagiae or Copenhageni serovars, while the C67 (64% DC) dog and A42 (60% DC) water isolates were not related (< 73.7% DC) to any of the 200 reference serovars; the closest serovars were the Leptospira noguchii Nicaragua and Orleans serovars, respectively.
Conclusion: This was the first molecular characterization of Colombian Leptospira spp isolates; these isolates will be used to develop a Colombian diagnostic panel.
Key words: Leptospira ; electrophoresis, gel, pulsed-field; Colombia
doi: http://dx.doi.org/10.7705/biomedica.v33i0.731
Caracterización molecular de serovariedades de Leptospira spp. aisladas de muestras de animales y agua en Colombia
Introducción. La leptospirosis es una infección bacteriana transmitida directa o indirectamente de animales a humanos, la cual puede resultar en una enfermedad hemorrágica grave, hepática o renal y pulmonar. Hay 20 especies de Leptospira conocidas y cientos de serovariedades, algunas de las cuales pertenecen a diferentes especies. Es esencial identificar las serovariedades patógenas y sus reservorios potenciales para enfocar estrategias de control.
Objetivo. Caracterizar las serovariedades de Leptospira aisladas de muestras de roedores, perros, cerdos y agua en Colombia.
Materiales y métodos. Las cepas de leptospiras aisladas fueron identificadas como patógenas usando la reacción en cadena de la polimerasa (PRC). Sus ADN y el ADN de Salmonella Braenderup H9812 (marcador de peso molecular) fueron cortados con NotI y corridos en electroforesis de campo pulsado. Los patrones de la ECP se analizaron con base en los criterios de tipificación para cepas bacterianas y el coeficiente de Dice, cuando se compararon con 200 cepas aisladas en otras partes del mundo. Los perfiles de ADN con un coeficiente de Dice entre 73,7 % y 100 % se consideraron pertenecientes a la misma especie.
Resultados. Todos los aislamientos fueron cepas patógenas y cinco se caracterizaron genéticamente. El aislamiento P275 (coeficiente de Dice: 84 %) y el P282 (coeficiente de Dice: 95 %) de cerdos, se relacionaron con Leptospira interrogans de serovariedad Pomona; el aislamiento de rata (I15) fue indistinguible de Leptospira interrogans de serovariedades Icterohaemorrhagiae o Copenhageni (coeficiente de Dice: 100 %), mientras que los aislamientos de perro (C67) y agua (A42) no se relacionaron (coeficiente de Dice <73,7 %) con ninguna de las 200 cepas de referencia; las más cercanas fueron Leptospira noguchii de serovariedades Nicaragua (coeficiente de Dice: 63 %) y Orleans (coeficiente de Dice: 60 %).
Conclusiones. Esta fue la primera caracterización molecular de serotipos de aislamientos colombianos, los cuales serían los primeros miembros de un panel diagnóstico colombiano.
Palabras clave. Leptospira , electroforesis en gel de campo pulsado, Colombia
doi: http://dx.doi.org/10.7705/biomedica.v33i0.731
Pathogenic Leptospira spp. are chronically maintained in the renal tubules of a wide range of wild and domestic mammals, and enzootic cycles are maintained by direct contact with infected urine or indirect contact with contaminated soil or water (1). More than 260 pathogenic serovars have been serologically identified and grouped into 24 serogroups adapted to different animal species (2). Although some animal species may act as maintenance hosts for some serovars, they can also be incidental hosts for other serovars, which may result in a range of clinical symptoms depending on the infecting strain, the geographical location and the host immune response (3). Leptospirosisis is a rural and occupational disease (4) considered to be an emerging zoonotic disease. In tropical and subtropical areas, the transmission of leptospires is increased during heavy rainfall, flooding, in conditions of poor sanitation and in areas of high host biodiversities; leptospirosis has therefore become a major public health problem in these areas (5).
The microscopic agglutination test (MAT) is the gold standard diagnostic assay for identifying leptospirosis but requires a large panel of live Leptospira serogroups/serovars to be maintained and used. Because it is difficult to isolate Leptospira serovars, the sensitivity of the MAT may be reduced when local serogroups/serovars are not included in these panels, thus limiting the possibility of characterizing other serovars (6). Knowledge of the prevalent serovars and their maintenance hosts and the monitoring of the emergence of new serovars is essential for understanding the epidemiology of the disease in a region and for developing effective control strategies (7).
Leptospira spp. isolates were identified as belonging to the pathogenic L. interrogans and the saprophytic L. biflexa species and were typed to the serovar level based on the expression of surface-exposed lipopolysaccharide (LPS) antigens using the cross agglutinin absorption test (CAAT) (8). Subsequently, molecular techniques such as DNA-DNA hybridization, restriction fragment length polymorphism (RFLP), and 16S rRNA sequence analysis (9) allowed the Leptospira genus to be classified into 14 pathogenic and intermediately pathogenic and 6 saprophytic species (10); however, a particular serovar can also belong to different species (11). Because of the epidemiological importance of precisely typing Leptospira organisms, molecular techniques such as multi-locus sequencing typing (MLST) (12) and pulsed-field gel eletrophoresis (PFGE) (13) are useful tools for genetically typing isolates at the serovar level. However, MLST cannot always be used for all Leptospira spp. as some serovars share the same sequence type or a particular serovar has multiple sequence types. In contrast, PFGE is generally applicable to all pathogenic species as this tool can identify strains of serovars that belong to different species and new serovars (10). In this study, we isolated and PFGE-characterized pathogenic Leptospira spp. isolates from animals (dogs, pigs and rats) and water samples collected in Colombia.
Materials and methods
Origin of Leptospira spp. isolates
The Leptospira spp. isolates were obtained from urine samples from two dogs (C67 and C76) (3.7%) of the 54 dogs sampled and from three pigs (P237, P275 and P282) (0.8%) of the 383 pigs sampled. Two other isolates (A42 and A44) (3.7%) were obtained from two of the 54 water samples. These isolates were obtained in 2008 from six pig farms in Monteria (Cordoba). One other isolate (I15) (6.3%) was obtained from a homogenized kidney sample from one of the 16 black rats ( Rattus rattus ) sampled from Barranquilla (Atlántico) in 2009. All of these isolates were maintained in three 5 ml culture tubes containing Ellinghausen-McCullough-Johnson-Harris (EMJH) liquid medium (Difco, USA) at 28°C.
Detection of pathogenic Leptospira spp isolates using a conventional polymerase chain reaction (PCR)
After five days of incubation, one 5-ml culture from each sample was centrifuged at 5000 x g for 5 min; the supernatants were then discarded and the pellets were re-suspended in 1 ml of media. DNA extraction was performed on 100 µl samples using Qiamp DNA mini kits (Qiagen, Hilden, Germany). The PCRs were performed in total volumes of 25 µl that contained 2-5 µl of DNA, 0.25 mM dNTPs (Bioline, USA), 1.0 IU of Taq DNA polymerase (Fermentas, Thermo Scientific, USA), 3.0 mM MgCl 2 , and 0.1 µM of LipL32/270F and LipL32/692R primers. The PCR program consisted of an initial cycle at 95°C for 5 minutes followed by 35 cycles at 94°C for 1 min and 55°C for 1 min, and a final extension step at 72°C for 5 min (14).
Leptospira isolate typing
The Leptospira isolates were grown in EMJH medium until a turbidity of 0.40 to 0.42 was reached (Dade Behring turbidity meter, Germany). Pulsed-field gel electrophoresis (PFGE) was performed as previously described (10). For this method, Leptospira DNA samples and Salmonella Braenderup H9812 strain DNA (used as standard size markers) were digested with Not I (Roche, Mannheim, Germany). PFGE was performed on a 1% agarose gel using a CHEF DR III (Bio-Rad) for 18 hr at 14°C, with 0.5 times Tris-Borate/EDTA (TBE) buffer recirculation, 2.16 and 35.07 second switching times at 120 o , a gradient of 6 V/cm, and a linear ramping factor. The resultant gel was stained with GelRed (Biotium, USA) for 30 minutes and analyses were performed using BioNumerics version 4.0 software (Applied Maths, Inc., Austin, TX, USA). The PFGE profiles were then compared with a library of over 200 Leptospira spp. reference serovars. In the PFGE gel analysis, genetic relatedness was based on the numbers of bands; the Dice coefficients were determined using BioNumerics version 4.0. Genetic relatedness was based on bacterial strain typing criteria and classified as indistinguishable (same number of bands with the same apparent size), closely related (two- or three-band differences), possibly related (four- to six-band differences) or unrelated (seven or more band differences) as previously described (15), and serovars with Dice coefficients between 73.7-100% were considered to be the same serovar.
Results
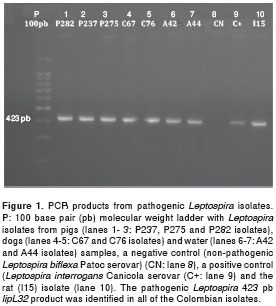
The 423 bp lipL32 gene fragment, specific to pathogenic Leptospira , was amplified by PCR in all of the pig (P237, P275 and P282), dog (C67, C76), and water samples (A42, A44) from the same farm in Monteria (Cordoba), and from the rat (I15) collected in Barranquilla (figure 1).
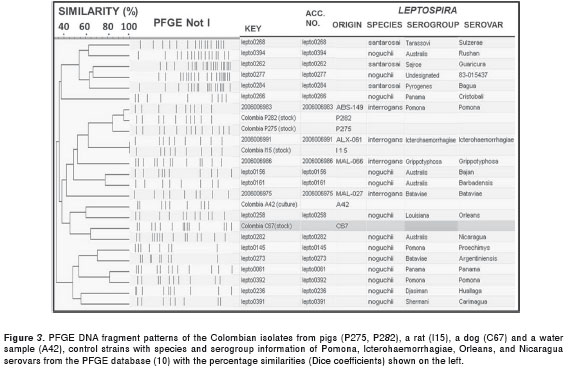
The PFGE analyses were only performed on isolates that attained turbidities of at least 0.40, and the numbers of PFGE bands obtained for each of the five isolates were as follows: P275 (n = 13), P282 (n = 10), C67 (n =14), A42 (n = 9) and I15 (n = 9) (figure 2). When the DNA profiles of the two pig isolates (P275 and P282) were analyzed, the most similar restriction band patterns were observed for the 2006006986 pathogenic Leptospira strain that belonged to the Leptospira interrogans Pomona serovar. The P275 isolate had 4 additional bands and an 84% DC and was therefore related to the Pomona serovar (figure 3). The P282 isolate, however, had only one additional band, resulting in a 95% DC and was therefore classified as more closely related to the Pomona serovar. In contrast, an indistinguishable restriction band pattern (100% DC) was obtained between the I15 rat isolate and the 200600691 strain, which belonged to the Leptospira interrogans Icterohaemorrhagiae serovar (figure 3). Although the C67 dog isolate had the same number of bands (n= 14) as lepto0282 Leptospira noguchii Nicaragua serovar, it had different restriction band patterns, resulting in a low DC of 64%. The A42 water isolate had two fewer bands than the nearest lepto0258 Leptospira noguchii Orleans serovar, which also resulted in a low DC of 60% (figure 3). Thus, because these two isolates had DCs below the threshold range (73.7-100%), they were classified as new serovars.

Discussion
To identify and understand alterations in leptospirosis epidemiology, it is essential to characterize the serovars of the circulating Leptospira strains in each endemic area so that appropriate control strategies can be implemented. In most endemic areas, this characterization has not been performed due to the slow growth of Leptospira organisms, requirements for special nutrients in the culture medium, frequent contamination with other micro-organisms, and the high cost of maintenance and genetic analyses (16). Although we obtained eight Leptospira isolates, PFGE could only be performed on five (5/8: 62.5%) of them. More efficient isolation and maintenance methods therefore need to be developed.
While the L. interrogans Icterohaemorrhagiae and Copenhageni serovars cause no disease in their rat reservoir hosts, they cause severe icteric Weil's disease in humans, which is particularly common in urban slum areas (17). This was consistent with the fact that the L. interrogans Icterohaemorrhagiae/Copenhageni serovar was isolated from a rat ( Rattus rattus ) (I15) that had been trapped in a poor urban human leptospirosis hot-spot' area of Barranquilla (18). The Leptospira noguchii serovars have been isolated from multiple animal species in Argentina, Nicaragua, Barbados, the United States of America and neighboring Panama and Peru, and from humans in the latter three countries (8,19). Interestingly, this serovar was also isolated from two humans (one of whom had severe icteric disease) who both had had contact with dogs and rats and from a dog with a fatal case of leptospirosis in Brazil (20). These results therefore demonstrate the putative importance of canine transmission of this serovar and its capacity to cause severe human disease.
This was the first report to type serovars of Colombian Leptospira isolates; the results showed that the Icterohaemorrhagiae/Copenhageni serovars that cannot be differentiated by PFGE analyses (11,21), the L. interrogans Pomona serovar, and possibly the L. noguchii Nicaragua and Orleans serovars are circulating in the Caribbean coast of Colombia. Despite the fact that a relatively low number of isolates were studied due to the well-documented difficulties in isolating and maintaining Leptospira spp., the isolate from one pig (P275) was found to have a different band pattern (84% Dice coefficient (DC)) than the reference Leptospira interrogans Pomona serovar, while the isolates from a dog (C67) (64% DC) and water (A42) (60% DC) were uniquely different from the closest reference Leptospira noguchii Nicaragua and Orleans serovars. These important isolates are the first Colombian isolates to be PFGE characterized; they will be further genetically analyzed using assays such as the gold standard cross agglutinin absorption test (CAAT) (9). Because it is advisable to use locally isolated Leptospira strains to obtain optimal sensitivities in serological assays (such as the MAT) (1), these uniquely different isolates will be used to establish a Colombian panel of genetically typed strains to improve Leptospira diagnosis in Colombia.
Acknowledgements
We thank the Dirección de Investigaciones, Desarrollo e Innovación de la Universidad del Norte, and the Centro de Investigaciones Universidad de Córdoba, código del proyecto FMV-2006.
Conflict of interest
The authors declare that no conflicts of interest exist.
Financial support
This study was supported by the Departamento Administrativo de Ciencia Tecnología e Innovación, Colciencias ID Project Number 1215-408-20551, Universidad del Norte, internal project 2011 Leptospirosis and the Universidad de Córdoba Project Number FMV-2006.
Corresponding author: Claudia M. Romero-Vivas, Grupo de Investigaciones en Enfermedades Tropicales, Universidad del Norte, km 5 vía a Puerto Colombia, Barranquilla, Colombia. Teléfono (575) 350 9478; fax (575) 350 9233 clromero@uninorte.edu.co
References
1. World Health Organization. Human leptospirosis: Guidance for diagnosis, surveillance and control. Geneva: WHO; 2003. p. 122. [ Links ]
2. Lau CL, Skelly C, Smythe LD, Craig SB, Weinstein P. Emergence of new leptospiral serovars in American Samoa-ascertainment or ecological change? BMC Infect Dis. 2012;12:19. http://dx.doi.org/10.1186/1471-2334-12-19 [ Links ]
3. Sykes JE, Hartmann K, Lunn KF, Moore GE, Stoddard RA, Goldstein RE. 2010 ACVIM small animal consensus statement on leptospirosis: Diagnosis, epidemiology, treatment, and prevention. J Vet Intern Med. 2011;25:1-13. http://dx.doi.org/10.1111/j.1939-1676.2010.0654.x [ Links ]
4. Everard CO, Maude GH, Hayes RJ. Leptospiral infection: A household serosurvey in urban and rural communities in Barbados and Trinidad. Ann Trop Med Parasitol. 1990;84:255-66. [ Links ]
5. Reis RB, Ribeiro GS, Felzemburgh RD, Santana FS, Mohr S, Melendez AX, et al . Impact of environment and social gradient on Leptospira infection in urban slums. PLoS Negl Trop Dis. 2008;2:e228. http://dx.doi.org/10.1371/journal.pntd.0000228 [ Links ]
6. Hull-Jackson C, Glass MB, Ari MD, Bragg SL, Branch SL, Whittington CU, et al . Evaluation of a commercial latex agglutination assay for serological diagnosis of leptospirosis. J Clin Microbiol. 2006;44:1853-5. http://dx.doi.org/10.1128/JCM.44.5.1853-1855.2006 [ Links ]
7. Levett PN. Leptospirosis. Clin Microbiol. 2001; 14:296-326. http://dx.doi.org/10.1128/CMR.14.2.296-326.2001 [ Links ]
8. Faine S, Adler B, Bolin C, Perolat P. Leptospira and leptos-pirosis. Second edition. Melbourne: MediSci; 1999. p. 57-63. [ Links ]
9. Ahmed N , Devi SM , Valverde ML , Vijayachari P , Machang'u RS , Ellis WA , et al . Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann Clin Microbiol Antimicrob. 2006;5:28. http://dx.doi.org/10.1186/1476-0711-5-28 [ Links ]
10. Galloway RL, Levett PN. Application and validation of PFGE for serovar identification of Leptospira clinical isolates. Plos Negl Trop Dis. 2010;14:e824. http://dx.doi.org/10.1371/journal.pntd.0000824 [ Links ]
11. De la Peña-Moctezuma A, Bulach DM, Kalambaheti T, Adler B. Comparative analysis of the LPS biosynthetic loci of the genetic subtypes of serovar Hardjo: Leptospira interrogans subtype Hardjoprajitno and Leptospira borgpetersenii subtype Hardjobovis. FEMS Microbiol Lett. 1999;177:319-26. http://dx.doi.org/10.1016/S0378-1097(99)00333-X [ Links ]
12. Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis. 2007;1:e56. http://dx.doi.org/10.1371/journal.pntd.0000056 [ Links ]
13. Herrmann JL, Bellenger E, Perolat P, Baranton G, Saint Girons I. Pulsed-field gel electrophoresis of Not I digests of leptospiral DNA: A new rapid method of serovar identification. J Clin Microbiol. 1992;30:1696-702. [ Links ]
14. Agudelo-Flórez P, Londoño AF, Quiroz VH, Ángel JC, Moreno N, Loaiza ET, et al . Prevalence of Leptospira spp. in urban rodents from a groceries trade center of Medellin, Colombia. Am J Trop Med Hyg. 2009;1:906-10. http://dx.doi.org/10.4269/ajtmh.2009.09-0195 [ Links ]
15. Tenover FC , Arbeit RD , Goering RV , Mickelsen PA , Murray BE , Persing DH , et al . I nterpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233-9. [ Links ]
16. Tulsiani SM , Graham GC , Dohnt MF , Burns MA , Craig SB . Maximizing the chances of detecting pathogenic leptospires in mammals: The evaluation of field samples and a multi-sample-per-mammal, multi-test approach. Ann Trop Med Parasitol. 2011;105:145-62. http://dx.doi.org/10.1179/136485911X12899838683205 [ Links ]
17. Maciel EA , de Carvalho AL , Nascimento SF , de Matos RB , Gouveia EL , Reis MG , et al . Household transmission of Leptospira infection in urban slum communities. PLoS Negl Trop Dis . 2008;2:e154. http://dx.doi.org/10.1371/journal. pntd.0000154 [ Links ]
18. Romero-Vivas CM, Cuello-Pérez M, Agudelo-Florez PM, Thiry D, Levett PN, Falconar AK. Urban transmission of pathogenic Leptospira in a main tropical sea-port city. Am J Trop Med Hyg. 2013;88:178-83 http://dx.doi.org/10.4269/ajtmh.2012.12-0232 [ Links ]
19. Brenner DJ, Kaufmann AF, Sulzer KR, Steigerwalt AG, Rogers FC, Weyant RS. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol. 1999;49:839-58. http://dx.doi.org/10.1099/ 00207713-49-2-839 [ Links ]
20. Silva EF, Cerqueira GM, Seyffert N, Seixas FK, Hartwig DD, Athanazio DA, et al . Leptospira noguchii and human and animal leptospirosis, Southern Brazil. Emerg Infect Dis. 2009;15:621-3. http://dx.doi.org 10.3201/eid1504.071669 [ Links ]
21. Naigowit P, Charoenchai S, Biaklang M, Seena U, Wangroongsarb P. Identification of clinical isolates of Leptospira spp. by pulsed field gel electrophoresis and microscopic agglutination test. Southeast Asian J Trop Med Public Health. 2007;38:97-103. [ Links ]













