Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Biomédica
Print version ISSN 0120-4157
Biomédica vol.34 no.4 Bogotá Oct./Dec. 2014
https://doi.org/10.7705/biomedica.v34i4.2246
ARTÍCULO ORIGINAL
doi: http://dx.doi.org/10.7705/biomedica.v34i4.2246
1 Grupo de Investigación en Alimentación y Nutrición Humana, Universidad de Antioquia, Medellín, Colombia
2 Grupo de Investigación GENMOL, Universidad de Antioquia, Medellín, Colombia
3 Grupo de Investigación GRICAFDE, Universidad de Antioquia, Medellín, Colombia
4 Grupo Vidarium, Centro de Investigación en Nutrición, Salud y Bienestar, Grupo Empresarial Nutresa, Medellín, Colombia
Author´s contributions:
Ana C. Orozco and Angélica M. Muñoz participated in the study design, the fieldwork, the genetic and environmental information analysis and the writing of the article.
Rosa M. Uscátegui conducted the anthropometric component of the study.
María V. Parra conducted the genetic component.
Fredy A. Patiño conducted the physical-activity component.
Luz M. Manjarrés conducted the food-consumption component.
Beatriz E. Parra led the biochemical component.
Alejandro Estrada led the statistical analysis.
Gloria M. Agudelo participated in the study design, in research management and data analysis.
Claudia M. Velásquez directed the research, designed the protocol and managed the logistics and statistical analysis.
All authors contributed in the analysis and the writing of the article.
Recibido: 31/01/14; aceptado: 04/06/14
Introduction : Obesity results from interaction between genetic and environmental risk factors.
Objective: To evaluate the effect of three gene variants and environmental factors on obesity and overweight in young people aged 10 to 18 years in a Colombian population.
Materials and methods: A total of 424 subjects were selected and separated into three groups for a cross-sectional study; 100 obese and 112 overweight subjects were matched with 212 normal-weight controls. Associations were evaluated between excess weight and three genetic polymorphisms ( UCP3- rs1800849, FTO -rs17817449, and CAPN10 -rs3842570), as well as the family history, the time spent watching television and playing video games, and the diet.
Results: A family history of obesity, the time spent watching television and playing video games, the lack of breastfeeding, a low consumption of cereals, legumes, fruits, vegetables, and a high consumption of fast foods were characteristics typically found in obese individuals compared to controls. A significant association between genotype I/I (SNP19 of CAPN10 ) and excess weight was found even with an active lifestyle. In addition, significant associations between the C/C genotype of the UCP3 gene and the G/G and T/T genotypes of the FTO gene and excess weight were found only in young sedentary individuals.
Conclusions: In this population, inadequate diet and sedentary lifestyle increased the risk of excess weight. Genotype I/I of SNP19 in CAPN10 was significantly associated with excess weight. In contrast, FTO and UCP3 variants exhibited effects only in sedentary environments.
Key words: Obesity, adolescent, risk factors, motor activity, Colombia.
doi: http://dx.doi.org/10.7705/biomedica.v34i4.2246
Una variante del gen CAPN10 y los factores ambientales muestran asociación con el exceso de peso en jóvenes colombianos.
Introducción. La obesidad resulta de la interacción entre factores de riesgo genéticos y ambientales.
Objetivo. Evaluar el efecto de tres variantes genéticas y factores ambientales en el exceso de peso en jóvenes de 10 a 18 años de Medellín, Colombia.
Materiales y métodos. Se hizo un estudio transversal en 424 jóvenes divididos en tres grupos: 100 obesos, 112 jóvenes con sobrepeso, y, pareados con ellos, 212 jóvenes con peso adecuado, que conformaron el grupo de control. Se evaluó la asociación entre tres polimorfismos genéticos ( UCP3 -rs1800849, FTO -rs17817449 y CAPN10 -rs3842570) y el exceso de peso, así como su interacción con antecedentes familiares de enfermedad, el tiempo dedicado a ver televisión y a jugar videojuegos y el consumo de alimentos.
Resultados. Los antecedentes familiares de obesidad, la dedicación de más de dos horas al día a ver televisión y jugar videojuegos, la falta de lactancia materna, el bajo consumo de cereales, legumbres, frutas y verduras y el gran consumo de comidas rápidas fueron más frecuentes entre los obesos que en los controles. Se observó una asociación significativa entre el genotipo I/I (SNP19 del CAPN10 ) y el exceso de peso, incluso en los jóvenes que llevaban una vida activa. Además, se encontró una asociación significativa entre los genotipos C/C del UCP3 y G/G y T/T del FTO y el exceso de peso, pero solo en los jóvenes sedentarios.
Conclusiones. En esta población, la alimentación inadecuada y el sedentarismo aumentaron el riesgo de exceso de peso. El genotipo I/I de SNP19 del CAPN10 se asoció significativamente con el exceso de peso. Algunas variantes del FTO y el UCP3 mostraron tener efecto solo en jóvenes sedentarios.
Palabras clave: obesidad, adolescente, factores de riesgo, actividad motora, Colombia.
doi: http://dx.doi.org/10.7705/biomedica.v34i4.2246
Obesity among children and teenagers has grown at an alarming rate in recent years and is increasingly beginning at younger ages. By 2010, there were 42 million overweight children worldwide and 35 million of them lived in developing countries (1). Notably, the Third National Health and Nutrition Examination Survey of the United States (NHANES III) showed that the number of obese children and teenagers had tripled between 1980 and 2010 (2). In Colombia, the situation is also alarming, as the percentage of teenagers with excess weight increased from 10.3% in 2005 (3) to 17.5% in 2010 (4).
Storing excess body fat negatively affects health, more so when accumulation by adipocytes exceeds the expansion capacity of cells, which promotes an inflammatory state that causes insulin resistance (IR). In addition, when lipids are stored in "unsuitable" organs, such as the pancreas, liver, and in muscles, this can cause lipotoxicity (5). Obesity increases the risk of cardiovascular disease, type 2 diabetes mellitus (T2DM), dyslipidemia, and hypertension . Obesity occurs when energy intake is greater than energy expenditure. Energy intake depends on the availability of foods as well as appetite control, which is a specific trait of individuals. Energy expenditure is a function of the amount of physical activity and energy metabolism efficiency. Appetite regulation and energy metabolism efficiency depend on gene variants involved in hunger, satiety, energy metabolism, and thermoregulation (6).
Increased obesity is related to the availability of foods of high caloric density, larger portions, and frequent fast-food consumption (7). The environmental component with the largest impact on energy expenditure is physical activity. Particularly for children and teenagers, physical activity includes time spent practicing sports, which contrasts with time spent on sedentary activities, such as playing video games and watching television (8).
Studies in twins and adopted children show evidence of a genetic predisposition to obesity with heritability ranging from 40% to 80% depending on age (9). Obesity acquired by complex inheritance results from the interaction of many genes with one another, as well as with environmental factors. The complex nature of obesity has been confirmed in 61 genome wide scans, which identified 127 obesity candidate genes and related phenotypes (10). However, these results vary greatly according to the ancestral origin of the populations studied.
In this study, we evaluated the fat mass and obesity associated ( FTO ) gene because it showed up as a candidate gene of body mass index in children and adults of different populations (11-13). The obesity-associated single nucleotide polymorphisms (SNP) are located in intron 1 of the FTO , all of which fall in a region of strong linkage disequilibrium (14); FTO encodes a protein that repairs alkylated DNA and RNA by oxidative demethylation (15); it has been implicated in increasing energy intake by regulating the expression of the genes that control appetite, such as leptin, leptin receptor, and neuropeptide Y (16). FTO has been associated with the modification of the degree of obesity in response to physical activity (17).
Further, we evaluated genetic variants in the uncoupling protei n-3 ( UCP3 ) and calpain-10 ( CAPN10) , because there is evidence that they are associated with obesity in American populations (18,19). UCP3 activity uncouples the hydrogen ion gradient in the respiratory chain to produce adenosine triphosphate (ATP), which increases metabolism and releases heat. UCP3 is homologous to the uncoupling protein–1 ( UCP1) and is also believed to be involved in thermoregulation (20). It has been suggested that UCP3 transports protonated fatty acids across the inner membrane, decreasing triglycerides and increasing oxidation and energy expenditure (21,22) . Polymorphism -55 C/T in the UCP3 has been associated with body mass index (BMI) in French Caucasians in interaction with physical activity (23) and body composition in Hispanic and non-Hispanic white females (24). This polymorphism is potentially interesting because it is located only six (6) bp from the TATA box, leading to the hypothesis that it could modify UCP3 gene expression and, therefore, modulate energy homeostasis and body weight regulation.
CAPN10 belongs to a superfamily of 15 cysteine proteases that are dependent on intracellular calcium. In this gene, SNPs 19, 43 and 63 have been implicated in decreased energy expenditure in several populations (25,26 ) . Among the mechanisms in the CAPN10 variants that lead to decreased energy expenditure are mitochondria-dependent negative regulation of glucose metabolism, decreased insulin secretion, and b 3 -adrenoceptor lipolytic sensitivity (27). In the Mexican pediatric population, a reduction in the CAPN-10 mRNA expression was observed in the excess weight groups with respect to the healthy weight group associated with InDel-19 (28).
Studying genotype x environment interactions in admixed populations gives a unique opportunity to find risk factors associated with overweight and obesity. Latin American populations, such as Colombians, are excellent examples of admixed populations since their genetic makeup consists of a recent mixture in different proportions of three ancestral populations: Amerindian, African, and European (29,30). The evaluation comprised environmental factors and genetic variants in the FTO, UCP3 and CAPN10 genes with respect to energy consumption and expenditure in normal weight, overweight and obese youngsters of Medellín, Colombia, to assess genotype x environment relations determining overweight in this population.
Materials and methods
Design
Four hundred twenty four (424) young people of both genders, ranging from 10 to 18 years of age, were selected from a previous study in which we investigated the prevalence of obesity and overweight among 1,060 young people from a company that provides health services. The BMI (weight/ height in Kg/m 2 ) was calculated, and participants were classified according to the 2007 World Health Organization (WHO) classification (31 ); all young people who exhibited excess weight were included for this study, both obese (BMI>percentile 98) (n=100) and overweight (p85<BMI<p98) (n=112). From the remaining youngsters with normal weight (p15<BMI<p85), 212 were selected and matched with the subjects according to age, gender, pubertal maturation, schooling level, birthplace of parents and grandparents, and social status.
We excluded from the study young people receiving medication or suffering from an illness that affected body composition, metabolism, or kidney function, or that hampered data collection; highly trained athletes who take part in competitive sports with high requirements of performance, and pregnant or lactating girls.
After reading and signing the informed consent form, the young people and their parents or guardians were asked about their geographical birthplace, perinatal data, family history of obesity, and socioeconomic status. After that, an 8 mL blood sample was taken for DNA extraction. Food consumption, physical activity, and the number of hours spent watching television and playing video games were assessed. In addition, infor mation on their pubertal maturation status and anthropometric measurem ents, such as weight (W), height (H), subscapular (SSF) and tricipital skin folds (TSF), and waist circumference (WC), were obtained to calculate the BMI and body fat percentage (BF%) indicators.
Anthropometry
Weight, height, and waist circumference were measured according to international standards. The Lohman equation was used to calculate the BF% (32). WC was considered high when > p90 according to the Third National Health and Nutrition Examination Survey of the United States (NHANES III) (33).
Pubertal maturation
Tanner´s classification (34,35) was used.
Food consumption
The 24-recall method was used. The information was collected at random in the subjects´ households throughout the week by trained nutrition students accompanied by the persons who prepared the food. Standardized food models were used (36).
Physical activity
The three (3)-day Physical Activity Recall questionnaire (3DPAR) (37) was used. Based on the average metabolic equivalent task (MET) minute/day, physical activity was classified as sedentary (1.0 to 1.4), indolent (1.4 to 1.6), active (1.6 to 1.9) and very active (1.9 to 2.5) (38).
Hours spent watching television and playing video games
The number of hours per day spent in these activ ities was obtained by questioning. Less than two hours a day indicated an active lifestyle, whereas more than two hours a day indicated a sedentary lifestyle (39).
Molecular data
DNA was extracted from the peripheral blood sample b y a standard method of salting out. Genotyping of rs1800849 (-55 C/T) and rs17817449 (C/G) were performed by polymerase chain reaction and restriction fragment length polymorphism (PCR- RFLP), and rs3842570 (IN/DEL 19) in CAPN10 was characterized using the size of the amplification product. The primers were designed on the basis of the DNA sequence of the target region accepted by the Genebank using the oligonucleotide design tool Primer 5.0 software. Forward (F) and reverse (R) primers were: 5´-GAGACTATATTAAAGCACC CCGGGTCAAGAGGAC -3´ and 5´- TCTGCTGC TTCTGGCTTGGCACTGGTCTTATACACCC -3´(UCP3-rs1800849); 5´- AGGACCTCCTATTTG GGACA -3´ and 5´- AGCTTCCATGGCTAGCATTA -3´ ( FTO -rs17817449), and 5´- GTTT GGTTCTCT TCAGCGTGGAG -3´ and 5´- ATGAACCCTGGCAG GGTCTAAG -3´ ( CAPN10 -rs3842570); 10 µl of PCR product generated from rs1800849 (-55 C/T) and rs17817449 (C/G) were digested with SmaI and AlwNI, respectively. The genotypes were resolved by 2.5% agarose electrophoresis stained with ethidium bromide (EtBr).
Ethical issues
In all cases, the researchers complied with Reso lution 8430 (1993) issued by the Colombian Ministry of Social Protection. In addition, this study was approved by the Bioethics Committee of the University of Antioquia Research Center (CBEIM- SIU). The consent form signed by the young people and their parents or guardians included the Declaration of Helsinki on ethical principles for medical research on human beings.
Statistical analysis
The descriptive statistics were analyzed with the Statistical Package for the Social Sciences, SPSS ® v19.0. The comparison of the qualitative environmental variables among the three weight groups was performed using c 2 tests. The residue normality assumption and homogeneity of variance were checked for variables using the Kolmogorov- Smirnov and Levene tests, respectively. The Kruskal-Wallis test was used to compare weight groups for variables with non-normal distribution of residues and heterogeneous variance, and the Mann-Whitney U test was used for multiple com parisons between weight groups. Spearman´s Rho correlation was used for variable correlation.
The University of Antioquia´s Dietary Intake Assessment Program v4 EVINDI was used to calculate the grams of food eaten by each individual. The Genpop v3.1 program (40) was used to calculate genotypic and allelic frequencies, and the Hardy- Weinberg equilibrium was tested for each weight group and the total sample. Co-dominant, dominant and recessive models were tested. Single-locus test of association between either SNP allele frequencies or SNP genotype frequencies and case-control status were carried out via standard contingency c 2 tests. We performed 10,000 per mutations for significance testing to determine empirical significance using PLINK (v1.07) (41). Logistic-regression models were fitted to estimate the odds ratio (OR) and 95% confidence intervals (CI); multiple logistic-regression models were also used to control for the effect of covariates, as well as to test the interaction between the SNP and an obesogenic environment, such as watching television and playing video games more than two hours/day, which is straightforward way to measure indirectly. Significance was considered at the level of p<0.05.
Results
No significant differences were detected among the groups with respect to the variables that matched ; statistical differences regarding the m easurements of anthropometric, personal and family history and physical activity among the groups are presented in table 1 .
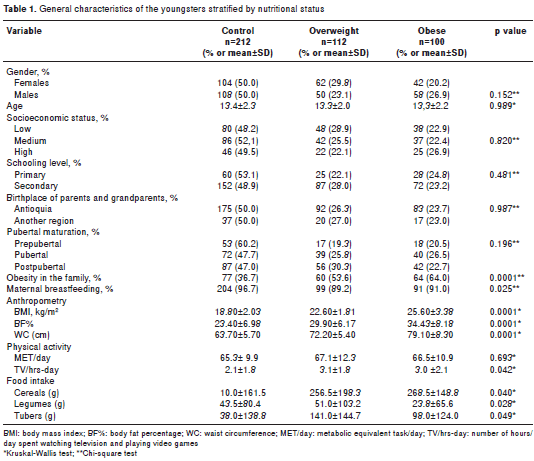
It is worth noting that all young people with healthy weight exhibited WC < p75. In contrast, 68.1% of subjects in the obese group and 8% in the over-weight group exhibited WC > p75 . A strong correlation was found between BMI and WC (Rho=0.909; p<0.0001).
A family history of obesity was significantly more prevalent in the obese and overweight groups compared to the control group. In contrast, subjects in the control group were breastfed in a significantly higher percentage than in the excess weight groups.
The physical activity level, measured in METs/ day, exhibited no significant difference among the groups. The obese and overweight groups spent significantly more hours per day watching television and playing v ideo games than the control group ( p=0.042). The overweight and obese groups con sumed significantly less cereals (p=0.040), fewer tubers and bananas (p=0.049) and legumes (p=0.028) than the normal-weight group.
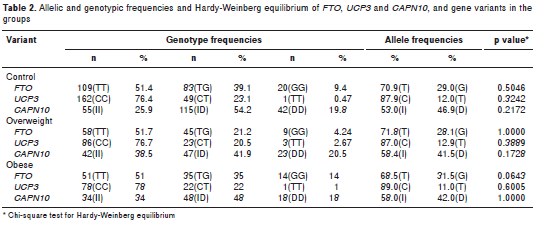
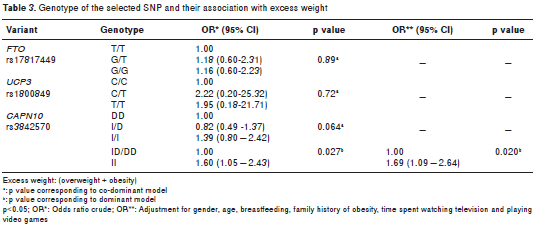
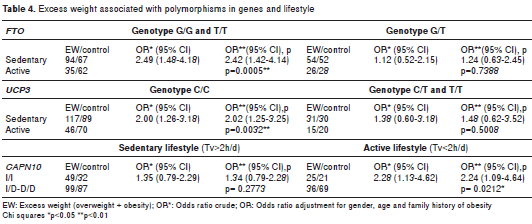
The genetic analysis revealed that the observed genotypic frequencies for each of the three poly morphisms were distributed according to the Hardy- Weinberg law (table 2). Allelic associations with excess weight were not significant for any of the three genetic variants (table 2). Only SNP19 of CAPN10 was associated with excess weight under the recessive model. We applied a permutation test to control the rate of error; these results maintained statistical significance, following adjustment for gender, age, breastfeeding, family history of obesity, and time spent watching television and playing video games (OR=1.69; CI 1.09-2.64, p=0.020) (table 3). Interaction between the SNPs and a sedentary lifestyle with excess weight was evaluated in this study. The interaction between the SNPs and sedentary lifestyle did not reach statistical significance (p>0.05). However, in a sub- group of 258 youth with G/G or T/T genotype of FTO , association was found with overweight in those with a sedentary lifestyle compared to those with an active lifestyle ( OR=2.42, CI 1.42-4.14, p=0.0005). Similarly, in 322 subjects with the C/C genotype of UCP3 , those with a sedentary lifestyle were associated with excess weight (OR=2.02; CI 1.25-3.25; p=0.0032). Finally, even in 151 young people with an active lifestyle, those with genotype I/I of SNP19 CAPN10 were associated to excess weight (OR=2.24; CI 1.09-4.64, p=0.0212) compared to genotypes I/D or D/D (table 4).
Discussion
This study found association between a family history of obesity and excess weight, which is possibly the result of the heritability. Obesity heritability was calculated by comparing the concordance between monozygotic (70% to 90%) and dizygotic (35% to 45%) twins in previous studies (42). The heritability and prevalence of obesity are influenced by age; thus, susceptibility genes may also be different in children and adults. Of 25 variants found in 13 adult obesity genes, only 15 variants found in nine loci were reported in children; the latter included the FTO gene studied here (43,44).
In contrast with previous reports, in this study the rs17817449 polymorphism (T/G) of FTO showed no allele or genotype association with obesity or overweight. However, our confidence intervals are quite wide, indicating that we have found an absence of evidence but no evidence of absence. Both the variant evaluated here and the one reported in Mexico were in complete linkage disequilibrium with the variant rs9939609 common in Europe (11,13). A meta-analysis of genome-wide association studies (GWAS ) in Asian population showed a strong effect of the G allele on BMI ( b =8.46%, SE=0.79, p=4.6 x 10 -27 ) (12). When the G allele frequency in this study (0.294) was compared with the frequencies obtained for European (0.447) and Mexican mestizo (0.211) populations (11,13), we were able to hypothesize that the ancestral state of the G allele is European because the genetic makeup of Colombian populations, evaluated by means of uniparental and biparental DNA markers, consists of 78% European, 16% Amerindian, and 6% African ancestry (45). There are many reasons for heterogeneity in genetic association studies; ethnic differences in genetic structure may produce different linkage disequilibrium (LD), thereby producing differences in the significance of the association test. Besides, differences in environmental, dietary or behavioral factors may also partially explain the heterogeneity in the genetic associations across ethnicities.
FTO plays a role in the regulation of genes involved in appetite and lipid metabolism, which can be modulated by environmental factors (46). It is well known that the association of FTO variants with BMI is observed more frequently in sedentary teenagers (16,17,47). In this population, the FTO gene showed an association in young people to the obesogenic factor of the time spent watching television and playing video games, as has occurred in other populations (17,43,48). We found a significant association between BMI and sedentary lifestyle, even after adjustment for physical activity. These results are consistent with previous studies (39,49).
Regarding UCP3 rs 1800849 (-55C/T), the frequency of the C allele was 0.88, which is consistent with the frequency reported by Franco, et al. , for an adult population of the same region of our study (0.86) despite the difference in sample size, 994 (50). This polymorphism (-55C/T) showed no allele or genotype association with obesity or overweight. UCP3 has not been reported in any GWAS, possibly because the frequencies of the risk variants of this gene are highly variable among populations. The associations reported in several Latin American populations were with type 2 diabetes mellitus (51) and were only found in association with obesity in American and European populations when consid ering the effects of variants in the adjacent gene UCP 2 (18,19,22,52) . Ochoa, et al. , studied a sample of 143 obese children and 170 controls in Spain and evaluated two additional variants in UCP2 (-866G/A and 45bp I/D) with the same results of this study (-55C/T): The frequency of the C allele was 0.85 in Spain and 0.88 in Colombia (50,52). These results might suggest that the ancestral state of the C allele in Colombia may be Spanish, as conquistadors conquered and colonized this region.
Consistent with our results, there was no association between obesity and genotypes of UCP3 in Spain; however, a significant association was observed between obesity and the haplotype -866 A, 45D, -55T of UCP3 (50,52). In our study, the association of the C/C genotype of UCP3 with excess weight was only found in sedentary teenagers. This outcome suggests that the expression of this gene in the study population could be mediated by physical activity, as other studies have shown (20-22).
The relationship between genetic variants of FTO and UCP3 with o verweight in young people with a sedentary lifestyle agrees with the finding that physical activity is highly beneficial, to the extent that it could be proposed as a replacement for drug therapies in diseases of the metabolic syndrome (39,53).
Few results have associated the CAPN10 gene variant rs3842570 with obesity or type 2 diabetes mellitus. However, rs 3842570 belongs to the diplotype 112/121 formed by the 43-19-63 SNPs identified as being of high risk for type 2 diabetes mellitus in the Mexican-American, Pima, Finnish, and German populations (26). The I allele frequency is highly variable across the continents, so the frequency found in this study (0.56) is lower than that reported in a Spanish population (0.64) (54 ), but similar to the frequencies reported for the Mexican-American (0.57), Hispanic (0.54) (55), and Arab Tunisian ( 0.57) (56 ) populations, while the frequency in Asian populations is 0.30 (25,56).
SNP19 of CAPN10 is located in an intron. This strongly suggests that it corresponds to a neutral variant. The particular associations that have been observed are the result of LD with one or more variants that increase or decrease the CAPN10 gene expression, whereby this allele can be a risk factor in one population and a protector factor in another (18,25,54,56). It has been proposed that LD can occur with variants in the CAPN10 gene promoter, which may involve changes in gene expression caused by environmental factors. In this study, genotype I/I was significantly associated with excess weight more than in the normal weight group under a recessive model, similar to data previously reported in Mexican children for the same age ranges (28); however, the association has little significance and should be studied in more detail. Interestingly, the association between excess weight and genotype I/I persisted even in those young people with an active lifestyle.
This study did not demonstrate genotype x envi ronment interaction, possibly because the effect of each variant on excess weight is very small ; also in chronic conditions, such as obesity, association only manifests itself later in life, after the accumulation of fat over time, due to an obesogenic environment. Although our study helps to understand excess weight in the population studied, it does have some limitations. It is a non-longitudinal study, the sample size is small and the associations are not very significant. Nevertheless, it constitutes the first approach to assess environmental and genetic risk factors associated with obesity in children and teenagers in a Colombian population. This and similar studies give us elements that help understand and prevent diseases associated with obesity. Furthermore, evaluation of SNPs in genetically mixed populations contributes to validate the universality of genetic markers for obesity predisposition.
In summary, our results concerning genotype and environment relations playing a role in obesity demonstrate the complex nature behind excess weight in an admixed population. On the one hand, we showed an association that suggested the relationship of genetics with obesity, as illustrated by genotype I/I of SNP19 of CAPN10, while, on the other hand, we identified the fact that low- fiber food and high fast-food intake, together with a sedentary lifestyle, are critical environmental variables contributing to the obesity epidemic in young people.
We wish to thank EPS SURA for allowing us to work with young users of their health service. We would like to thank Juan S. Escobar for improvements in the manuscript.
The authors declare that there are no conflicts of interest.
This work was financed by Colciencias Contract 203 (2010) and the University of Antioquia 2011-2012 Sustainability Strategy.
Corresponding author: Claudia María Velásquez, Sede de Investigación Universitaria, Universidad de Antioquia, Calle 62 N° 52-59, laboratorio 413, Medellín, Colombia Teléfonos: (574) 219 6497, 219 6498
claudia.velasquez@udea.edu.co y claudiamariavelasquez@gmail.com
1. World Health Organization. Global strategy on diet, physical activity and health: Childhood overweight and obesity. Fecha de consulta: 25 de enero de 2013. Disponible en: http://www.who.int/dietphysicalactivity/childhood/es. [ Links ]
2. Ryan AS, Roche AF, Kuczmarski RJ. Weight, stature, and body mass index data for Mexican Americans from the third national health and nutrition examination survey (NHANES III, 1988-1994). Am J Hum Biol. 1999;11:673-86. http://dx.doi.org/10.1002/(SICI)1520-6300(199909/10)11:5%3C673::AID- AJHB10%3E3.3.CO;2-9 [ Links ]
3. ICBF, Profamilia, Instituto Nacional de Salud, Universidad de Antioquia, OPS. Encuesta Nacional de la Situación Nutricional en Colombia (ENSIN)-2005. Bogotá, D.C.: ICBF; 2005. [ Links ]
4. Instituto Colombiano de Bienestar Familiar, Profamilia, Instituto Nacional de Salud, Universidad de Antioquia, Organización Panamericana de la Salud. Encuesta Nacional de la Situación Nutricional en Colombia 2010. Bogotá, D.C.: Panamericana Formas e Impresos S.A.; 2011. [ Links ]
5. Pietilainen KH, Rog T, Seppanen-Laakso T, Virtue S, Gopalacharyulu P, Tang J, et al . Association of lipidome remodeling in the adipocyte membrane with acquired obesity in humans. PLoS Biol. 2011;9:e1000623. http://dx.doi.org/10.1371/journal.pbio.1000623 [ Links ]
6. Pankov YA. Genetic variations in energy balance regulation. Biomed Khim. 2010;56:152-67. http://dx.doi.org/10.1134/S1990750810030017 [ Links ]
7. Elbel B. Consumer estimation of recommended and actual calories at fast food restaurants. Obesity (Silver Spring). 2011;19:1971-8. http://dx.doi.org/10.1038/oby.2011.214 [ Links ]
8. Davison KK, Marshall SJ, Birch LL. Cross-sectional and longitudinal associations between TV viewing and girls´ body mass index, overweight status, and percentage of body fat. J Pediatr. 2006;149:32-7. http://dx.doi.org/10.1016/j.jpeds.2006.02.003 [ Links ]
9. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al . Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937-48. http://dx.doi.org/10.1038/ng.686 [ Links ]
10. Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, et al . The human obesity gene map: The 2005 update. Obesity (Silver Spring). 2006;14:529- 644. http://dx.doi.org/10.1038/oby.2006.71 [ Links ]
11. Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, et al. Genome- wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18-24. http://dx.doi.org/10.1038/ng.274 [ Links ]
12. Xi B, Wang C, Wu L, Zhang M, Shen Y, Zhao X, et al . Influence of physical inactivity on associations between single nucleotide polymorphisms and genetic predisposition to childhood obesity. Am J Epidemiol. 2011;173:1256-62. http://dx.doi.org/10.1093/aje/kwr008 [ Links ]
13. Villalobos-Comparán M, Flores-Dorantes M, Villarreal- Molina M, Rodríguez-Cruz M, García-Ulloa AC, Robles L, et al. The FTO gene is associated with adulthood obesity in the Mexican population. Obesity (Silver Spring). 2008;16:2296-301. http://dx.doi.org/10.1038/oby.2008.367 [ Links ]
14. The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789-96. http://dx.doi. org/10.1038/nature02168 [ Links ]
15. Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469-72. http://dx.doi.org/10.1126/science.1151710 [ Links ]
16. Fawcett KA, Barroso I. The genetics of obesity: FTO leads the way. Trends Genet. 2011;26:266-74. http://dx.doi.org/10.1016/j.tig.2010.02.006 [ Links ]
17. Kilpelainen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, et al . Physical activity attenuates the influence of FTO variants on obesity risk: A meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8:e1001116. http://dx.doi.org/10.1371/journal.pmed.1001116 [ Links ]
18. Liu YJ, Liu PY, Long J, Lu Y, Elze L, Recker RR, et al. Linkage and association analyses of the UCP3 gene with obesity phenotypes in Caucasian families. Physiol Genomics. 2005;22:197-203. http://dx.doi.org/10.1152/physiolgenomics.00031.2005 [ Links ]
19. Alonso A, Martí A, Corbalán MS, Martínez-González MA, Forga L, Martínez JA. Association of UCP3 gene -55C>T polymorphism and obesity in a Spanish population. Ann Nutr Metab. 2005;49:183-8. http://dx.doi.org/10.1159/000086883 [ Links ]
20. Hancock AM, Clark VJ, Qian Y, Di Rienzo A. Population genetic analysis of the uncoupling proteins supports a role for UCP3 in human cold resistance. Mol Biol Evol. 2010;28:601-14. http://dx.doi.org/10.1093/molbev/msq228 [ Links ]
21. Chan CB, Kashemsant N. Regulation of insulin secretion by uncoupling protein. Biochem Soc Trans. 2006;34:802-5. http://dx.doi.org/10.1042/BST0340802 [ Links ]
22. Jia JJ, Zhang X, Ge CR, Jois M. The polymorphisms of UCP2 and UCP3 genes associated with fat metabolism, obesity and diabetes. Obes Rev. 2009;10:519-26. http://dx.doi.org/10.1111/j.1467-789X.2009.00569.x [ Links ]
23. Otabe S, Clement K, Dina C, Pelloux V, Guy-Grand B, Froguel P, et al . A genetic variation in the 5´ flanking region of the UCP3 gene is associated with body mass index in humans in interaction with physical activity. Diabetologia. 2000;43:245-9. http://dx.doi.org/10.1007/s001250050037 [ Links ]
24. Damcott CM, Feingold E, Moffett SP, Barmada MM, Marshall JA, Hamman RF, et al. Genetic variation in uncoupling protein 3 is associated with dietary intake and body composition in females. Metabolism. 2004;53:458-64. http://dx.doi.org/10.1016/j.metabol.2003.11.019 [ Links ]
25. Harris F, Biswas S, Singh J, Dennison S, Phoenix DA. Calpains and their multiple roles in diabetes mellitus. Ann N Y Acad Sci. 2006;1084:452-80. http://dx.doi.org/10.1196/annals.1372.011 [ Links ]
26. Pihlajamaki J, Salmenniemi U, Vanttinen M, Ruotsalainen E, Kuusisto J, Vauhkonen I, et al . Common polymorphisms of calpain-10 are associated with abdominal obesity in subjects at high risk of type 2 diabetes. Diabetologia. 2006;49:1560-6. http://dx.doi.org/10.1007/s00125-006-0270-z [ Links ]
27. Carlsson E, Fredriksson J, Groop L, Ridderstrale M. Variation in the calpain-10 gene is associated with elevated triglyceride levels and reduced adipose tissue messenger ribonucleic acid expression in obese Swedish subjects. J Clin Endocrinol Metab. 2004;89:3601-5. http://dx.doi.org/10.1210/jc.2003-032105 [ Links ]
28. Mendoza-Lorenzo P, Salazar AM, Cortés-Arenas E, Saucedo R, Taja-Chayeb L, Flores-Dorantes M, et al . The reduction of calpain-10 expression is associated with risk polymorphisms in obese children. Gene. 2013;516:126-31. http://dx.doi.org/10.1016/j.gene.2012.12.053 [ Links ]
29. Rojas W, Parra MV, Campo O, Caro MA, Lopera JG, Arias W, et al. Genetic make-up and structure of Colombian populations by means of uniparental and biparental DNA markers. Am J Phys Anthropol. 2010;143:13-20. http://dx.doi.org/10.1002/ajpa.21270 [ Links ]
30. Galanter JM, Fernández-López JC, Gignoux CR, Barnholtz-Sloan J, Fernández-Rozadilla C, Via M, et al. Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. PLoS Genet. 2012;8:e1002554. http://dx.doi.org/10.1371/journal.pgen.1002554 [ Links ]
31. de Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ. 2007;85:660-7. http://dx.doi.org/10.2471/BLT.07.043497 [ Links ]
32. Lohman T, Roche A, Martorell R. Anthropometric stand ardization reference manual. Champaing: Human Kinetics Books; 1988. 177 p. [ Links ]
33. Fernández JR, Redden DT, Pietrobelli A, Allison DB. Waist circumference percentiles in nationally representative samples of African- American, European-American, and Mexican-American children and adolescents. J Pediatr. 2004; 145:439-44. http://dx.doi.org/10.1016/j.jpeds.2004.06.044 [ Links ]
34. Marshall WA, Tanner JM. Variations in pattern of pubertal changes in girls. Arch Dis Child. 1969;44:291-303. http://dx.doi.org/10.1136/adc.44.235.291 [ Links ]
35. Marshall WA, Tanner JM. Variations in the pattern of pubertal changes in boys. Arch Dis Child. 1970;45:13-23. http://dx.doi.org/10.1136/adc.45.239.13 [ Links ]
36. Manjarrés L. Métodos para precisar la recolección de la ingesta dietética en estudios poblacionales. Perspect Nutr Hum. 2007;9:155-63. [ Links ]
37. Pate RR, Ross R, Dowda M, Trost SG. Validation of a 3-day physical activity recall instrument in female youth. Pediatric Exercise Science. 2003;15:257-565. [ Links ]
38. Ainsworth BE, Haskell WL, Whitt MC, Irwin ML, Swartz AM, Strath SJ, et al. Compendium of physical activities: An update of activity codes and MET intensities. Med Sci Sports Exerc. 2000;32:S498-504. http://dx.doi.org/10.1097/00005768-200009001-00009 [ Links ]
39. Gómez LF, Parra DC, Lobelo F, Samper B, Moreno J, Jacoby E, et al . Television viewing and its association with overweight in Colombian children: Results from the 2005 National Nutrition Survey: A cross sectional study. Int J Behav Nutr Phys Act. 2007;4:41. http://dx.doi.org/10.1186/1479-5868-4-41 [ Links ]
40. Raymon M, Rousset F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248-9. [ Links ]
41. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A tool set for whole- genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559-75. http://dx.doi.org/10.1086/519795 [ Links ]
42. Guerra C, Vila J, Apolinaire J, Cabrera A, Santana I, Almaguer P. Factores de riesgo asociados a sobrepeso y obesidad en adolescentes. Medisur. 2009;7:25-34. [ Links ]
43. Field AE, Aneja P, Austin SB, Shrier LA, de Moor C, Gordon-Larsen P, et al . Race and gender differences in the association of dieting and gains in BMI among young adults. Obesity (Silver Spring). 2007;15:456-64. http://dx.doi.org/10.1038/oby.2007.560 [ Links ]
44. Zhao J, Bradfield JP, Li M, Wang K, Zhang H, Kim CE, et al . The role of obesity-associated loci identified in genome-wide association studies in the determination of pediatric BMI. Obesity (Silver Spring). 2009;17:2254-7. http://dx.doi.org/10.1038/oby.2009.159 [ Links ]
45. Johnson NA, Coram MA, Shriver MD, Romieu I, Barsh GS, London SJ, et al. Ancestral components of admixed genomes in a Mexican cohort. PLoS Genet. 2011;7: e 1002410. http://dx.doi.org/10.1371/journal.pgen.1002410 [ Links ]
46. Sovio U, Mook-Kanamori DO, Warrington NM, Lawrence R, Briollais L, Palmer CN, et al. Association between common variation at the FTO locus and changes in body mass index from infancy to late childhood: The complex nature of genetic association through growth and development. PLoS Genet. 2011;7:e1001307. http://dx.doi.org/10.1371/journal.pgen.1001307 [ Links ]
47. Demerath EW, Lutsey PL, Monda KL, Linda WH, Bressler J, Pankow JS, et al. Interaction of FTO and physical activity level on adiposity in African-American and European- American adults: The ARIC study. Obesity (Silver Spring). 2011;19:1866-72. http://dx.doi.org/10.1038/oby.2011.131 [ Links ]
48. Vimaleswaran KS, Li S, Zhao JH, Luan J, Bingham SA, Khaw KT, et al. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am J Clin Nutr. 2009;90:425-8. http://dx.doi.org/10.3945/ajcn.2009.27652 [ Links ]
49. Mendoza JA, Zimmerman FJ, Christakis DA. Television viewing, computer use, obesity, and adiposity in US preschool children. Int J Behav Nutr Phys Act. 2007;4:44. http://dx.doi.org/10.1186/1479-5868-4-44 [ Links ]
50. Franco-Hincapié L, Duque CE, Parra MV, Gallego N, Villegas A, Ruiz-Linares A, et al. Association between polymorphism in uncoupling proteins and type 2 diabetes in a northwestern Colombian population. Biomédica. 2009; 29:108-18. http://dx.doi.org/10.7705/biomedica.v29i1.46 [ Links ]
51. Comuzzie AG, Almasy L, Cole SA, Boss O, Giacobino JP, Muzzin P, et al. Linkage exclusion analysis of the chromosome 11 region containing UCP2 and UCP3 with obesity-related phenotypes in Mexican Americans. Int J Obes Relat Metab Disord. 2000;24:1065-8. http://dx.doi.org/10.1038/sj.ijo.0801257 [ Links ]
52. Ochoa MC, Santos JL, Azcona C, Moreno-Aliaga MJ, Martínez-González MA, Martínez JA, et al. Association between obesity and insulin resistance with UCP2-UCP3 gene variants in Spanish children and adolescents. Mol Genet Metab. 2007;92:351-8. http://dx.doi.org/10.1016/j.ymgme.2007.07.011 [ Links ]
53. Múnera NE, Uscátegui RM, Parra BE, Manjarrés LM, Patiño F, Velásquez CM, et al. Factores de riesgo ambientales y componentes del síndrome metabólico en adolescentes con exceso de peso. Biomédica. 2012;32:77-91. http://dx.doi.org/10.1590/S0120-41572012000100010 [ Links ]
54. Sáez ME, González-Sánchez JL, Ramírez-Lorca R, Martínez-Larrad MT, Zabena C, González A, et al . The CAPN10 gene is associated with insulin resistance phenotypes in the Spanish population. PLoS One. 2008;3:e2953. http://dx.doi.org/10.1371/journal.pone.0002953 [ Links ]
55. Song Y, You NC, Hsu YH, Sul J, Wang L, Tinker L, et al . Common genetic variation in calpain-10 gene ( CAPN10 ) and diabetes risk in a multi-ethnic cohort of American postmenopausal women. Hum Mol Genet. 2007;16:2960- 71. http://dx.doi.org/10.1093/hmg/ddm256 [ Links ]
56. Ezzidi I, Mtiraoui N, Nemr R, Kacem M, Al-Khateeb GM, Mahjoub T, et al. Variants within the calpain-10 gene and relationships with type 2 diabetes (T2DM) and T2DM-related traits among Tunisian Arabs. Diabetes Metab. 2011;36:357-62. http://dx.doi.org/10.1016/j.diabet.2010.03.005 [ Links ]













