Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Biomédica
Print version ISSN 0120-4157
Biomédica vol.34 supl.1 Bogotá Apr. 2014
https://doi.org/10.7705/biomedica.v34i0.1702
ARTÍCULO ORIGINAL
doi: http://dx.doi.org/10.7705/biomedica.v34i0.1702
1Grupo de Análisis Bioinformático, GABi, Centro de Investigación y Desarrollo en Biotecnología, CIDBIO, Bogotá, D.C., Colombia
2Laboratorio de Modificación de ARN y Enfermedades Mitocondriales, Centro de Investigación Príncipe Felipe, Valencia, España
3Grupo de Genética y Bioquímica de Microorganismos, GEBIOMIC, Universidad de Antioquia, Medellín, Colombia
Author contributions:
Alfonso Benítez-Páez and María Eugenia Armengod designed this study.
Alfonso Benítez-Páez, Sonia Cárdenas-Brito and Mauricio Corredor performed all sequence analyses and protein modelling.
Alfonso Benítez-Páez and Magda Villarroya performed experimental approaches such as directed mutagenesis, resistance assays, cell culture, protein techniques, and HPLC of modified nucleosides.
Recibido: 17/06/13; aceptado: 27/08/13
Introduction: Aminoglycosides like streptomycin are well-known for binding at specific regions of ribosome RNA and then acting as translation inhibitors. Nowadays, several pathogens have been detected to acquire an undefined strategy involving mutation at non structural ribosome genes like those acting as RNA methylases. rsmG is one of those genes which encodes an AdoMet-dependent methyltransferase responsible for the synthesis of m 7 G527 in the 530 loop of bacterial 16S rRNA. This loop is universally conserved, plays a key role in ribosomal accuracy, and is a target for streptomycin binding. Loss of the m 7 G527 modification confers low-level streptomycin resistance and may affect ribosomal functioning.
Objectives: After taking into account genetic information indicating that some clinical isolates of human pathogens show streptomycin resistance associated with mutations at rsmG , we decided to explore new hot spots for mutation capable of impairing the RsmG in vivo function and of promoting low-level streptomycin resistance.
Materials and methods: To gain insights into the molecular and genetic mechanism of acquiring this aminoglycoside resistance phenotype and the emergence of high-level streptomycin resistance in rsmG mutants, we mutated Escherichia coli rsmG and also performed a genotyping study on rpsL from several isolates showing the ability to grow at higher streptomycin concentrations than parental strains.
Results: We found that the mutations at rpsL were preferentially present in these mutants, and we observed a clear synergy between rsmG and rpsL genes to induce streptomycin resistance.
Conclusion: We contribute to understand a common mechanism that is probably transferable to other ribosome RNA methylase genes responsible for modifications at central sites for ribosome function.
Key words: RNA/biosynthesis, streptomycin, methylation, Escherichia coli , aminoglycosides, mutagenesis.
doi: http://dx.doi.org/10.7705/biomedica.v34i0.1702
Mutaciones en genes modificadores de ARN ribosómico y la resistencia a aminoglucósidos: el caso del gen rsmG
Introducción. Los aminoglucósidos son moléculas antibióticas capaces de inhibir la síntesis de proteínas bacterianas tras su unión al ribosoma procariota. La resistencia a aminoglucósidos está clásicamente asociada a mutaciones en genes estructurales del ribosoma bacteriano; sin embargo, varios estudios recientes han demostrado, de forma recurrente, la presencia de un nuevo mecanismo dependiente de mutación que no involucra genes estructurales. El gen rsmG es uno de ellos y se caracteriza por codificar una metiltransferasa que sintetiza el nucleósido m 7 G527 localizado en el loop 530 del ribosoma bacteriano, este último caracterizado como sitio preferencial al cual se une la estreptomicina.
Objetivo. Partiendo de las recientes asociaciones clínicas entre las mutaciones en el gen rsmG y la resistencia a estreptomicina, este estudio se propuso la caracterización de nuevos puntos calientes de mutación en este gen que puedan causar resistencia a estreptomicina usando Escherichia coli como modelo de estudio.
Materiales y métodos. Se indagó sobre el mecanismo genético y molecular por el cual se adquiere la resistencia a estreptomicina y su transición a la resistencia a altas dosis mediante mutagénesis dirigida del gen rsmG y genotipificación del gen rpsL .
Resultados. Se encontró que la mutación N39A en rsmG inactiva la proteína y se reportó un nuevo conjunto de mutaciones en rpsL que confieren resistencia a altas dosis de estreptomicina.
Conclusiones. Aunque los mecanismos genéticos subyacentes permanecen sin esclarecer, se concluyó que dichos patrones secuenciales de mutación podrían tener lugar en otros genes modificadores del ARN bacteriano debido a la conservación evolutiva y al papel crítico que juegan tales modificaciones en la síntesis de proteínas.
Palabras clave: ARN/biosíntesis, estreptomicina, metilación, K, aminoglucósidos, mutagénesis.
doi: http://dx.doi.org/10.7705/biomedica.v34i0.1702
Aminoglycosides are probably the most widely used group of broad-spectrum antibiotics to over-come bacterial infections and to prevent pathogen dissemination. Despite their antimicrobial therapy potential, toxic effects and physiology dysfunctions are well-known in patients under long-lasting or recurrent aminoglycoside treatment (1,2). In biochemical terms, aminoglycosides are composed of modified sugar moieties where several amine groups are present to confer cationic properties to the molecule as a whole. Consequently, amino- glycosides bind to certain RNA regions of prokaryotic ribosome RNA (rRNA) (3,4) and then they act as protein synthesis inhibitors.
Nowadays, mutations at non structural ribosome genes frequently appear to confer aminoglycoside resistance, including the genes encoding the different AdoMet-dependent methylases responsible for post-transcriptional modification of rRNAs. One of the classical and first reported cases is rsmA , where the presence of mutations inactivating the RsmA function allows kasugamycin resistance (5-10). RsmA is a well-studied dimethyltansferase enzyme which catalyzes the synthesis of the m 6 2 A ribonucleotide at positions 1618 and 1619 of 16S rRNA in E. coli . This family of proteins is widely distributed in all major kingdoms of life: Eukaryota, Archaea and Bacteria. Although methylations at rRNA are almost fully depicted, at least in Bacteria and Eukaryota cells, their evolutionary and functional meaning remain unclear. However, few studies have recently analyzed the role of methylations at 16S and 23S rRNA in maintaining ribosome accuracy, which is experimental evidence to support the function of rRNA methylations for the first time (11,12).
Among aminoglycosides, streptomycin is probably the best known given its antibiotic effect in tuberculosis treatment in the late 1940's. Strepto-mycin binds to 16S rRNA in four different domains, one including nucleotides G526 and G527 in the 530 loop, thus perturbing the A-site function and leading to translational misreading (13,14). Classically, streptomycin resistance mutations have been preferentially associated with mutations in 16S rRNA (530 loop, and helix 44 proximity comprising the 1402-1410 and 1490-1498 positions) and S12 protein ( rpsL ) (15-17).
Interestingly, recent reports indicate that mutations in a specific rRNA methyltransferase gene confer streptomycin resistance (18,19). The gene involved in this resistance pattern is rsmG (previously known as gidB ). rsmG encodes a Class-I and AdoMet-dependent RNA methyltransferase that specifically methylates the N7 position of the purine ring at G527 of 16S rRNA in E. coli . Loss of RsmG activity confers low-level streptomycin resistance and inexplicably prompts the appearance of high-level streptomycin resistance mutations (18-22). We recently reported an extensive mutational analysis to address the hot spots of mutation within the rsmG gene to confer streptomycin resistance (23). Simultaneously, we presented evidence for the critical role of RsmG activity of a certain set of residues located at the catalytic and co-factor binding sites of the enzyme (23).
The aim of this study was to explore the molecular genotype of the E. coli rsmG mutants explaining transition from low-level to high-level streptomycin resistance, and to present new hot spots of mutation to impair the RsmG function, all of which are likely genetic traits for screening in human pathogens and clinical isolates to explain potential streptomycin resistance phenotype.
Materials and methods
Bacterial strains, plasmids, and media
Escherichia coli rsmG and a parental wild-type strain were obtained from the National BioResource Project (NIG, Japan). The pBAD-TOPO and pET15b plasmids were obtained from Invitrogen® and Novagen®, respectively. C-terminal Flag- tagged rsmG was cloned into pBAD-TOPO using these respective forward and reverse primers: rsmG-flag-F 5'-GATACCATGGTGCTCAACAAAC TCTCCTTACTGC and rsmG-flag-R 5'-TTATTA tttgtcgtcgtcgtctttatagtc AATTTTATTTGCTTTAATCACCACC. C-terminal 6His-tagged rsmG was cloned into the pET15b plasmid using primers: rsmG-his-F 5'-GATA CCATGG TGCTCAACAAACTCTCCTTACTGC and rsmG-his-R 5'-AGATA GGATCC TTATTA atgatggtgatgatggtg AATTTTATTTGCTTTAATCAC CACC, where the Nco I and BamH I restriction sites were encoded (sequences in bold). The sequence encoding respective tags are denoted by underlined lower case. LBT (Luria Bertani broth containing 40 µg/ml thymine) and LAT (LBT containing 20 g of Difco agar per liter) were used for routine cultures and for the plating of E. coli . LBT or LAT media with 20 µg/ml or 100 µg/ml of streptomycin (Sigma) were used to test low-level and high-level streptomycin resistance, respectively.
Sequence and comparative structural analysis
The phylogenetic distribution of RsmG was studied by downloading a set of 134 sequences, belonging to the major eubacterial groups, from the Uniprot database (24). They were aligned using the Probcons program with 1,000 refinement iterations (25). Then, a Hidden Markov Model (HMM)-based profile was built using the HMMER v 3.0 application and default parameters (26). A consensus sequence of the RsmG family was extracted and used for massive searching in a local bacteria genome database consisting in more than 1,200 non redundant complete genomes downloaded from the NCBI FTP site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/). A general procedure for detecting functional residues in protein families based on sequence analysis was used (27). The RsmG protein from Bacillus subtilis (Protein Data Bank – PDB - id 1XDZ) was used as a template to model the missing loops of the E. coli RsmG protein (28) (PDB id 1JSX). The E. coli RsmG sequence was submitted to the SwissModel server (29) to be modeled over the 1XDZ template. Then, the best model based on ANOLEA and QMEAN energy minimization was used as a template for the manual docking of the AdoMet co-factor using the RsmG protein from Thermus thermophilus (20) (PDB id 3G88). The alpha carbons of the glycines conforming the canonical AdoMet-binding motif 73 GxGxG 77 were used to superimpose both structures with DeepView (30). The RSM deviation between the AdoMet motifs was calculated to be 0.14 Å. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, USA (31).
Protein techniques
Western blotting, in vivo complementation assays, AdoMet affinity and in vivo rRNA modification activity measuring of the RsmG wild-type and N39A mutant were performed as previously reported (23).
Emergence of spontaneous high-level streptomycin resistance and genotyping
The mutants exhibiting high-level streptomycin resistance were obtained from wild-type strains and their respective rsmG mutants. Serial dilutions of a defined cell suspension (5 ml) of each strain were spread on LAT plates containing 100 µg/ml of streptomycin (Sigma). Serial dilutions of the cell suspension were also plated on LAT plates without streptomycin to determine the numbers of viable cells in the cell suspension. The colonies showing high-level streptomycin resistance were isolated and passed twice in LB media containing 100 µg/ml of streptomycin. The genomic DNA from those isolates with stable high-level streptomycin resistance was isolated using the DNA Isolation Kit for Cells and Tissues (Roche), and the rpsL gene was amplified by PCR using the Expand High Fidelity PCR System (Roche) and the following primers: rpsL-F 5'-ACACCTTTTCGGCATCGCC and rpsL-R 5'-CTACGAGTTTAGTTTGACATTTAAG to hybridize ~100 nt upstream and downstream, respectively, of the rpsL coding region. Sanger sequencing from purified PCR was conducted using forward primer rpsL-F.
Results
Sequence analysis of the RsmG family of proteins
For the purpose of retrieving new residues that are critical for the RsmG function and the probable hot spot sites for mutation, an amino acid profile of the RsmG family of proteins was built from 134 representative sequences of major bacterial groups (see Materials and methods). Then, new residues were selected as hot spots according to their localization in the three-dimensional structure (holoenzyme modelled for E. coli RsmG protein) and the information content inferred from the Hidden Markov Model (HMM) analysis of the multiple sequence alignment (MSA), where those residues located close to the catalytic site of the enzyme were of interest. After discarding the previously characterized ones (23), we aimed to study a set of the residues lacking from available RsmG proteins structures (20,28) where some displayed high predominance (figure 1A).
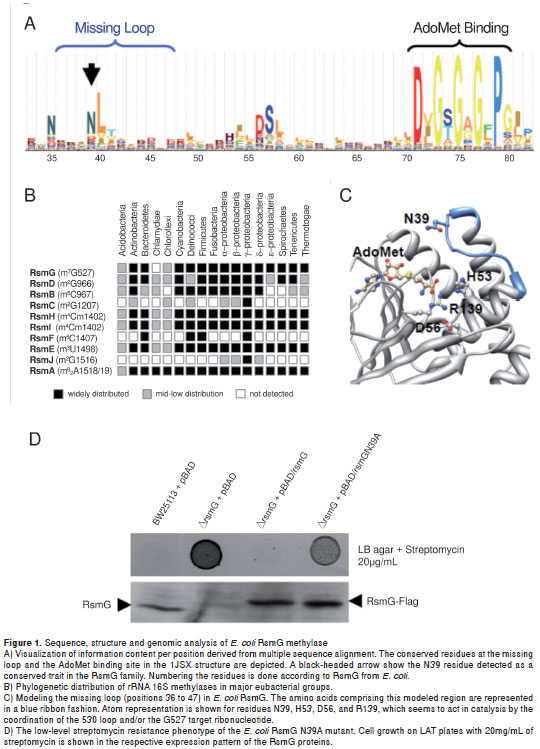
In the E.coli RsmG structure, the region comprising the amino acids from positions 36 to 47 was lacking. After loop modelling (see Materials and methods), this structural region was predicted to be located over the co-factor binding site (figure 1C), and N39 was predicted as being a critical residue given the conservation pattern along the alignment and the disposition of its side chain facing the active site of protein. Therefore, experimental approaches were designed to test the functional relevance of N39 in RsmG activity (see the next section). Moreover, we further analyzed the phylogenetic distribution of rsmG among bacterial species, as well as the occurrence profile of other RNA methyltransferases that modify E. coli 16S rRNA. This aim could provide information about its presence in bacterial genomes which could present or acquire streptomycin resistance features by the mutation of rsmG gene. We found that enzyme RsmG was encoded in most of the bacterial genomes analyzed (more than 1,200) with very few exceptions (figure 1B).
Accordingly, rsmG seems to play a critical role in the ribosome function of prokaryotes because its conservation pattern resembled those observed for RsmD, RsmH/RsmI, and RsmE, whose acting sites converged at the P site of the 30S ribosome subunit where codon-anticodon pairing occurs between the mRNA and aminoacylated-tRNA during elongation (32,33). Therefore, whereas the conserved methylations at positions G966, C1402, and 1498 have been demonstrated to be central for translation initiation (34,35), m 7 G527 could participate in tRNA selection in promoting the correct rotation of the 530 loop, thus maintaining the A-site proper function and avoiding translational misreading (13,14), hypothetical function that needs to be studied in deep given the growth phenotype of rsmG mutants (18).
Although only rsmG and rsmA , among the most conserved genes acting as RNA methylases of 16S rRNA, have been associated with aminoglycoside resistance, it would not be surprising to find aminoglycoside-resistant bacterial strains with point mutations of other very conserved genes such as rsmE, rsmI and rsmH, which act in the proximity of helix 44, a critical structure for translation initiation (36) and a selective site for aminoglycoside binding (37). In fact, recent reports have demonstrated that deletion of rsmF gene encoding the methylase acting on m 5 C1407 position of E. coli 16S rRNA, at helix 44 proximity, is also associated to low-level aminoglycoside resistance (38). Despite its low conservation level (figure 1B), this data reinforces our hypothesis to acquire resistance through inactivation of housekeeping genes involved in rRNA methylation at selective sites for aminoglycoside binding. Consequently, all of them, and the remaining set of unknown genes encoding uncharacterized and species specific rRNA modifying genes, must be considered potential markers for aminoglycoside resistance screening.
Functional characterization of N39 from E. coli RsmG
The coding region of E. coli rsmG was cloned in vectors pBAD and pET15b to evaluate the role of N39 within the protein. The N39 residue was mutated to alanine in both constructs, and the ability to confer streptomycin resistance, to modify rRNA and to bind AdoMet were explored for this mutant and wild type proteins. After transforming the ? rsmG cells with the pBAD plasmids encoding wild type and N39A versions of the C-terminal Flag-tagged RsmG protein, and being spotted on LB plates with a low streptomycin concentration (20 µg/mL), the cells carrying the N39A mutant of RsmG showed the original phenotype of the Δ rsmG cells, low-level streptomycin resistance (figure 1D), indicating that the N39A protein is inactive despite it being expressed (figure 1D). The activity of RsmG N39A in vivo was evaluated using the same growth conditions.
As a result, the level of the 16S rRNA modification was studied by measuring the m 7 G nucleoside accumulated in the Δ rsmG cells harboring wild type and N39A versions of the RsmG-Flag encoded in pBAD plasmids. When comparing the amount of m 7 G detected by HPLC after the hydrolysis of 16S rRNA from those strains expressing RsmG and the N39A mutant, we observed in the rsmG mutant carrying the N39A construct only a fraction of 0.24 ± 0.04 of m 7 G accumulated in the rsmG mutant carrying the plasmid-encoded wild type RsmG. This result indicates that mutation N39A largely affects the RsmG function, as previous inferred from the streptomycin resistance assay. To further explore whether the co-factor binding in RsmG is impaired with the N39A mutation, the Surface Plasmon Resonance (SPR) approach was used to measure AdoMet affinity. AdoMet affinity assays were performed by over-expressing the C-terminal 6His-tagged RsmG proteins encoded in the pET15b plasmids from E. coli BL21 (DE3) cells. The RsmG wild-type protein showed an affinity of 0.39 ± 0.02 µM by AdoMet, whereas the N39A mutant displayed a slight reduced (~3-fold) AdoMet affinity, being 1.19 ± 0.36 µM.
When comparing with a previous analysis, this difference in AdoMet affinity is not expected to largely influence RsmG activity given that an RsmG mutant G75A, which directly affects the AdoMet binding site, presents an almost four-fold loss of affinity and maintains the RsmG protein active given both its phenotype in the streptomycin resistance assay and the in vivo activity of the protein (23). Therefore, we speculate that the N39 residue could be involved in catalysis, maybe in helping to maintain the proper position of the 530 loop and/or specifically the G527 ribonucleotide at the active enzyme site.
Characterization of ΔrsmG cells with high-level streptomycin resistance
To further study the genotype associated with the high-level streptomycin resistance of rsmG mutants (18), we performed a selection assay to recover the rsmG mutants that are able to grow at high streptomycin concentrations (100 µg/ mL). After this assay, we recovered 42-fold more emergence frequency of high-level streptomycin resistance in Δ rsmG cells than in the wild type ones. This inexplicably high frequency for the appearance of high-level streptomycin resistance cells has been previously identified in both E. coli and Mycobacterium tuberculosis and, in some cases, they involve mutations in the S12-encoding gene, rpsL (18). Consequently, we decided to analyze the rpsL genotype in these emerged high-level streptomycin resistant isolates. A summary of this genotyping is shown in table 1. Globally, we found that 63% of these isolates had mutations in rpsL . In all, 16 point mutations were identified in 12 isolates, of which three showed double point mutations. The most frequent mutation was K88Q (7/16), reported to be found in high-level streptomycin resistant isolates (39). Despite a previous study reporting no profound effects of the K88Q mutation in translation accuracy (40), the variation in K88 seems central for this spontaneous drug resistant phenotype of rsmG mutants given that two other type K88R mutations were found, evidencing that the mutations at the K88 codon covered more than 55% of the mutations reported for rpsL in this study. Notably, we present a new set of mutations that confer a high-level streptomycin resistance phenotype to rsmG mutants by involving the L7, K44, H77, and P91 residues of the S12 ribosome protein.
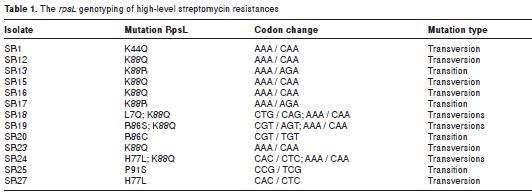
The transition/transversion ratio (R) observed (R = 0.397), which differed from the theoretical probability of occurrence (R = 0.5), together with the high prevalence of the rpsL mutations in combination with the rsmG mutation, support the notion of an obscure molecular strategy to promote streptomycin resistance and to evade drug therapy, as frequently found in clinical isolates; therefore, the complete understanding of these coupled mutations can help to design better strategies for microbial control and eradication.
Discussion
The present study aimed at shedding light on our understanding of new molecular mechanisms for aminoglycoside resistance. In addition to the large set of residues detected as hot spots of mutation in the E. coli rsmG gene (23), we present a new mutation in this gene, which largely impairs enzyme activity, and it could be useful to detect microorganisms with low-level streptomycin resistance that are potentially able to develop high-level resistance. Moreover, we present the partial genotyping of emerged high-level streptomycin resistant isolates by demonstrating a sequential mutation process to acquire such a phenotype.
Mutations that confer a high-level streptomycin resistance phenotype to rsmG mutants preferentially appear on rpsL . We present a new set of mutations apart from K88Q which confer the ability to E. coli to grow in media at high streptomycin concentrations. Initial low-level aminoglycoside resistance steps, involving mutations at rRNA methylation genes, followed by the emergence of high-level resistant mutants, seems a common feature for this recently characterized treatment-evading strategy of microbes and human pathogens (18,41).
Therefore, we can expect a similar behavior and mutation-acquiring mechanisms for other genes acting at critical sites for the ribosome decoding function, like those methylating ribonucleotides in the proximity of helix 44. The step-by-step point mutation-acquiring process can indicate that cells would suffer loss of cell fitness in the very early selection process under antibiotic treatment, which would help evade the drug therapy, but would cause drastically reduced growth in a competitive environment.
Nevertheless, fast adaptation mechanisms would be active in order to acquire compensatory mutations in specific genes, which would increase the level of antibiotic resistance in addition to attenuating the effect of the initial disadvantageous mutations. This likely short-term evolution mechanism for antibiotic resistance, based on the mutation of RNA methylation genes, must be studied in depth given that the complete understanding of this genetic response of pathogens against therapy could help us to design new strategies in order to avoid the emergence and dissemination of high-level aminoglycoside resistance. Indeed, this mechanism of mutation is not restricted to generate aminoglycoside resistance given that more recent reports have associated mutations at rRNA methylations genes with linezolid resistance in clinical isolates (42,43).
The authors declare no competing interest.
This study was supported by the Colombian Agency for Science, Technology and Innovation (Colciencias), grant 5817-5693-4856 to ABP, and the Spanish Ministry of Economy and Competitiveness (BFU2010-19737) to MEA.
Corresponding author: Alfonso Benítez-Páez, Calle 64A N° 52-53, interior 8, oficina 203, Bogotá, D.C., Colombia Tel: (57) (310) 559 0633 abenitez@cidbio.org
1. Fischel-Ghodsian N. Genetic factors in aminoglycoside toxicity. Pharmacogenomics. 2005;6:27-36 . http://dx.doi.org/10.1517/14622416.6.1.27 [ Links ]
2. I nciardi JF. Aminoglycoside toxicity. J Infect Dis. 1993;168: 1594-5. http://dx.doi.org/10.1093/infdis/168.6.1594 [ Links ]
3. Recht MI, Douthwaite S, Puglisi JD. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. Embo J. 1999;18:3133-8. http://dx.doi.org/10.1093/emboj/18.11.3133 [ Links ]
4. Recht MI, Fourmy D, Blanchard SC, Dahlquist KD, Puglisi JD. RNA sequence determinants for aminoglycoside binding to an A-site rRNA model oligonucleotide. J Mol Biol. 1996;262:421-36. http://dx.doi.org/10.1006/jmbi.1996.0526 [ Links ]
5. Sparling PF, Ikeya Y, Elliot D. Two genetic loci for resistance to kasugamycin in Escherichia coli . J Bacteriol. 1973;113:704-10. [ Links ]
6. Zimmermann RA, Ikeya Y, Sparling PF. Alteration of ribosomal protein S4 by mutation linked to kasugamycin-resistance in Escherichia coli . Proc Natl Acad Sci U S A. 1973;70:71-5. [ Links ]
7. Helser TL, Davies JE, Dahlberg JE. Mechanism of kasugamycin resistance in Escherichia coli . Nat New Biol. 1972;235:6-9. [ Links ]
8. Poldermans B, Goosen N, van Knippenberg PH. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli . I. The effect of kasugamycin on initiation of protein synthesis. J Biol Chem. 1979;254:9085-9. [ Links ]
9. Poldermans B, Roza L, van Knippenberg PH. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli . III. Purification and properties of the methylating enzyme and methylase-30 S interactions. J Biol Chem. 1979;254:9090-100. [ Links ]
10. Poldermans B, van Buul CP, van Knippenberg PH. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli . II. The effect of the absence of the methyl groups on initiation of protein biosynthesis. J Biol Chem. 1979;254:9090-3. [ Links ]
11. Benítez-Páez A, Villarroya M, Armengod ME. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA. 2012;18:1783-95. http://dx.doi.org/10.1261/rna.033266.112 [ Links ]
12. Kimura S, Suzuki T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010;38:1341-52. http://dx.doi.org/10.1093/nar/gkp1073 [ Links ]
13. Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340-8. http://dx.doi.org/10.1038/35030019 [ Links ]
14. Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129-77. http://dx.doi.org/10.1146/annurev.biochem.74.061903.155440 [ Links ]
15. Finken M, Kirschner P, Meier A, Wrede A, Bottger EC. Molecular basis of streptomycin resistance in Mycobacterium tuberculosis : Alterations of the ribosomal protein S12 gene and point mutations within a functional 16S ribosomal RNA pseudoknot. Mol Microbiol. 1993;9:1239-46. http://dx.doi.org/10.1111/j.1365-2958.1993.tb01253.x [ Links ]
16. Springer B, Kidan YG, Prammananan T, Ellrott K, Bottger EC, Sander P. Mechanisms of streptomycin resistance: Selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob Agents Chemother. 2001;45:2877-84. http://dx.doi.org/10.1128/AAC.45.10.2877-2884.2001 [ Links ]
17. Vila-Sanjurjo A, Lu Y, Aragonez JL, Starkweather RE, Sasikumar M, O'Connor M. Modulation of 16S rRNA function by ribosomal protein S12. Biochim Biophys Acta. 2007;1769:462-71. http://dx.doi.org/10.1016/j.bbaexp.2007.04.004 [ Links ]
18. Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, et al . Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol. 2007;63:1096-106. http://dx.doi.org/10.1111/j.1365-2958.2006.05585.x [ Links ]
19. Nishimura K, Hosaka T, Tokuyama S, Okamoto S, Ochi K. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J Bacteriol. 2007;189:3876-83. http://dx.doi.org/10.1128/JB.01776-06 [ Links ]
20. Gregory ST, Demirci H, Belardinelli R, Monshupanee T, Gualerzi C, Dahlberg AE, et al . Structural and functional studies of the Thermus thermophilus 16S rRNA methyltransferase RsmG. RNA. 2009;15:1693-704. http://dx.doi.org/10.1261/rna.1652709 [ Links ]
21. N ishimura K, Johansen SK, Inaoka T, Hosaka T, Tokuyama S, Tahara Y, et al . Identification of the RsmG methyltransferase target as 16S rRNA nucleotide G527 and characterization of Bacillus subtilis rsmG mutants. J Bacteriol. 2007;189:6068-73. http://dx.doi.org/10.1128/JB.00558-07 [ Links ]
22. Wong SY, Lee JS, Kwak HK, Via LE, Boshoff HI, Barry CE 3rd. Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis . Antimicrob Agents Chemother. 2011;55:2515-22 . http://dx.doi.org/10.1128/AAC. 01814-10 [ Links ]
23. Benítez-Páez A, Villarroya M, Armengod ME. Regulation of expression and catalytic activity of Escherichia coli RsmG methyltransferase. RNA. 2012;18:795-806. http://dx.doi.org/ 10.1261/rna.029868.111 [ Links ]
24. The Uniprot Consortium. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43-7. http://dx.doi.org/10.1093/nar/gks1068 [ Links ]
25. Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15:330-40. http://dx.doi.org/10.1101/gr.2821705 [ Links ]
26. Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755-63. http://dx.doi.org/10.1093/bioinformatics/14.9.755 [ Links ]
27. Benítez-Páez A, Cardenas-Brito S, Gutiérrez AJ. A practical guide for the computational selection of residues to be experimentally characterized in protein families. Brief Bioinform. 2012;13:329-36. http://dx.doi.org/110.1093/bib/bbr052 [ Links ]
28. Romanowski MJ, Bonanno JB, Burley SK. Crystal structure of the Escherichia coli glucose-inhibited division protein B (GidB) reveals a methyltransferase fold. Proteins. 2002;47:563-7. http://dx.doi.org/10.1002/prot.10121 [ Links ]
29. Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195-201. http://dx.doi.org/10.1093/bioinformatics/bti770 [ Links ]
30. Guex N, Peitsch MC. SWISS-MODEL and the Swiss- PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714-23. [ Links ]
31. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al . UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605-12. http://dx.doi.org/10.1002/jcc.20084 [ Links ]
32. Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065-77. http://dx.doi.org/10.1016/j.cell.2006.08.032 [ Links ]
33. Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, et al . Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935-42. http://dx.doi.org/10.1126/science.1131127 [ Links ]
34. Cunningham PR, Nurse K, Bakin A, Weitzmann CJ, Pflumm M, Ofengand J. Interaction between the two conserved single-stranded regions at the decoding site of small subunit ribosomal RNA is essential for ribosome function. Biochemistry. 1992;31:12012-22. [ Links ]
35. Cunningham PR, Nurse K, Weitzmann CJ, Ofengand J. Functional effects of base changes which further define the decoding center of Escherichia coli 16S ribosomal RNA: Mutation of C1404, G1405, C1496, G1497, and U1498. Biochemistry. 1993;32:7172-80. [ Links ]
36. Qin D, Liu Q, Devaraj A, Fredrick K. Role of helix 44 of 16S rRNA in the fidelity of translation initiation. RNA. 2012;18:485-95. http://dx.doi.org/10.1261/rna.031203.111 [ Links ]
37. Ogle JM, Brodersen DE, Clemons WM, Jr., Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897-902. http://dx.doi.org/10.1126/science.1060612 [ Links ]
38. Gutiérrez B, Escudero JA, San Millán A, Hidalgo L, Carrilero L, Ovejero CM, et al . Fitness cost and interference of Arm/Rmt aminoglycoside resistance with the RsmF housekeeping methyltransferases. Antimicrob Agents Chemother. 2012;56:2335-41. http://dx.doi.org/10.1128/AAC.06066-11 [ Links ]
39. Bottger EC, Springer B, Pletschette M, Sander P. Fitness of antibiotic-resistant microorganisms and compensatory mutations. Nat Med. 1998;4:1343-4. http://dx.doi.org/10.1038/3906 [ Links ]
40. Toivonen JM, Boocock MR, Jacobs HT. Modelling in Escherichia coli of mutations in mitoribosomal protein S12: Novel mutant phenotypes of rpsL. Mol Microbiol. 1999;31:1735-46. http://dx.doi.org/10.1046/j.1365-2958.1999.01307.x [ Links ]
41. Ochi K, Kim JY, Tanaka Y, Wang G, Masuda K, Nanamiya H, et al . Inactivation of KsgA, a 16S rRNA methyltransferase, causes vigorous emergence of mutants with high-level kasugamycin resistance. Antimicrob Agents Chemother. 2009;53:193-201. http://dx.doi.org/10.1128/AAC.00873-08 [ Links ]
42. Gao W, Chua K, Davies JK, Newton HJ, Seemann T, Harrison PF, et al . Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 2010;6:e1000944. http://dx.doi.org/10.1371/journal.ppat.1000944 [ Links ]
43. LaMarre JM, Howden BP, Mankin AS. Inactivation of the indigenous methyltransferase RlmN in Staphylococcus aureus increases linezolid resistance. Antimicrob Agents Chemother. 2011;55:2989-91. http://dx.doi.org/10.1128/ AAC.00183-11 [ Links ]













