Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Biomédica
versão impressa ISSN 0120-4157
Biomédica vol.35 no.1 Bogotá jan./mar. 2015
https://doi.org/10.7705/biomedica.v35i1.2276
ARTÍCULO ORIGINAL
doi: http://dx.doi.org/10.7705/biomedica.v35i1.2276
1 Departamento de Microbiologia, Inmunologia e Parasitologia, Faculdade de Ciências Médicas, Universidade do Estado do Rio de Janeiro, Rio de Janeiro, Brasil
2 Laboratório de Parasitologia Molecular, Instituto de Biofísica Carlos Chagas Filho, Universidade Federal do Rio, Rio de Janeiro, Brasil
3 Instituto Federal de Educação, Ciência e Tecnologia de Rio de Janeiro, Campus Maracanã, Laboratório de Genética Molecular, Rio de Janeiro, Brasil
Author´s contributions:
Verônica Dias Gonçalves and Françoise Bohrer Lengruber contributed equally to the manuscript through their dissertations, which had the guidance and cooperation of the other six authors.
Recibido: 27/02/14; aceptado: 19/11/14
Introduction: Multidrug-resistant Enterobacteriaceae, particularly those resistant to gentamicin, have become one of the most important causes of nosocomial infections.
Objective: We sought to investigate the presence of genes conferring resistance to aminoglycosides, specially to gentamicin, in Klebsiella pneumoniae and Escherichia coli multidrug-resistant strains isolated from different clinical materials among patients hospitalized in a university hospital in Rio de Janeiro, Brazil.
Materials and methods: Ten colonization strains and 20 infection strains were evaluated during three decades (1980 to 2010) using selective media containing 8 µg/ml of gentamicin. Thirty strains were tested for antimicrobial susceptibility. Twenty two strains were subjected to plasmid DNA extraction and 12 to hybridization assays using as probe a 1.9 kb plasmid DNA fragment from one of the K. pneumoniae strains isolated from faecal samples. This fragment was sequenced and assigned to the GQ422439 GenBank record. PCR was also performed using oligonucleotides designed for aminoglycoside-modifying enzymes.
Results: An accC2 acetylase, besides transposons and insertion sequences, were evidenced. Twenty-four (80%) of the isolates were positive for the aacC2 gene in agreement with antibiotic susceptibility testing profiles, indicating the persistent presence of this gene throughout the three decades. We detected high molecular weight plasmids in 54,5% of the strains. Of the tested strains, 91% showed positive signal in the hybridization assays.
Conclusion: A gene codifying for one specific aminoglycoside-modifying enzyme was detected all throughout the three decades. Our data back the adoption of preventive measures, such as a more conscious use of antimicrobial agents in hospital environments, which can contribute to control the dissemination of microorganisms harboring resistance gene plasmids.
Key words: Enterobacteriaceae, infection; drug resistance, multiple, bacterial; enzymes, aminoglycoside, plasmids, acetylesterase.
doi: http://dx.doi.org/10.7705/biomedica.v35i1.2276
Detección y caracterización de enterobacterias resistentes a múltiples fármacos, portadoras del gen modificador de aminoglucósidos en un hospital universitario de Río de Janeiro, Brasil, durante tres décadas
Introducción. Las enterobacterias resistentes a la gentamicina se asocian frecuentemente a infecciones hospitalarias.
Objetivo. Verificar la presencia de los genes que confieren resistencia a los aminoglucósidos, específicamente a la gentamicina, en cepas de Klebsiella pneumoniae y Escherichia coli multirresistentes, obtenidas de pacientes internados en un hospital universitario de Río de Janeiro.
Materiales y métodos. Se recolectaron y evaluaron 10 cepas de colonización y 20 de infección entre 1980 y 2010, utilizando medios selectivos enriquecidos con gentamicina (8 µg/ml). Se obtuvieron 30 cepas en las que se determinó la resistencia a los antibióticos por medios fenotípicos. Veintidós muestras se sometieron a extracción de ADN plasmídico y se hicieron ensayos de hibridización en 12 de ellas, usando como sonda un fragmento de ADN plasmídico de 1,9 kb obtenido de una cepa de K. pneumoniae aislada de muestra fecal. Este fragmento fue secuenciado y correspondió al registro GQ422439 del GenBank. Se verificó la presencia de genes de enzimas modificadoras de aminoglucósidos mediante reacción en cadena de la polimerasa.
Resultados. En las cepas analizadas se evidenció la presencia de la acetilasa accC2, además de transposones y secuencias de inserción. Veinticuatro aislamientos (80 %) fueron positivos para el gen aacC2 en concordancia con los perfiles de sensibilidad a los antibióticos, lo que indicó su persistencia a lo largo de las tres décadas. Se detectaron plásmidos de alto peso molecular en 54,5 % de las cepas. El 91 % de las cepas analizadas mostró signos positivos en las pruebas de hibridación.
Conclusión. Se detectó la persistencia de un gen codificador de una enzima modificadora de aminoglucósidos a lo largo de las tres décadas. Los resultados indican que las medidas de prevención, tales como un uso más responsable de los agentes antimicrobianos en el ambiente hospitalario, pueden contribuir al control de la diseminación de microorganismos que albergan plásmidos de genes de resistencia.
Palabras clave: Enterobacteriacae, infección, farmacorresistencia bacteriana múltiple, enzimas, aminoglicósidos, plásmidos, acetilesterasa.
doi: http://dx.doi.org/10.7705/biomedica.v35i1.2276
In the last decades, multidrug-resistance of nosocomial bacteria has been a concern for healthcare professionals around the world (1,2). Through gene acquisition or mutation, bacteria can become resistant to one or several antimicrobial drugs. Gene acquisition involving horizontal transfer is a more frequent resistance mechanism than mutations, especially among enterobacteria, and it involves plasmid, gene cassette and transposon participation (3).
Besides resistance genes, resistance plasmids can carry other genes that also contribute to the pathogenicity of microorganisms (4). The integrons can house several resistance genes in Gram-negative bacteria. These genes may be found within resistance cassettes, and their different combinations can be generated by site-specific recombination (5). Transposons are also important in establishing bacterial resistance genes. The capacity of these mobile genetic elements ´to circulate´ between plasmids and chromosomes is decisive for the dissemination of resistance-encoding genes to antimicrobial agents as beta-lactams and aminoglycosides (6,7).
Aminoglycosides are highly potent, broad-spectrum antibiotics, widely used for the treatment of life- threatening infections. Resistance to these drugs occurs through several mechanisms that can coexist in the same cell; nevertheless, these resistance is often due to enzymatic inactivation by acetyltransferases, nucleotidyltransferases (adenylyltransferases), and phosphotransferases. The accC2 gene is among the most frequently detected in strains of Enterobacteriaceae isolated from clinical samples (8). Many of these genes are associated with transposons, which help to the rapid dissemination of drug resistance across species boundaries (9).
In a previous study with Enterobacteriaceae strains of hospital origin we searched for genetic elements which express multidrug-resistance including resis tance to aminoglycosides (10). Probes were built to detect plasmids encoding these multidrug-resistance traits. In the present work, we sought to investigate the presence of multidrug-resistant bacteria harboring aminoglycosid e- resistanc e- encoding genes, particularly to gentamicin, in clinical especimens from a university hospital, and the persistence of these genetic elements during three decades. In another study, Gonçalves (11) detected the aacC2 gene in strains of hospital origin, showing how the gene sequence was flanked by transposons and insertion sequences.
Materials and methods
Isolated strains under study
We analyzed 30 Enterobacteriaceae strains (18 K. pneumoniae and 12 E. coli strains) from 28 patients interned in a 600-bed tertiary university hospital in Rio de Janeiro, Brazil. Strains isolated from stool and urine samples of two of the patients were used. The strains were randomly selected from the multidrug-resistant Enterobacteriaceae (gentamicin- resistant) collected in the university hospital. Both infection and colonization strains were isolated from samples recovered in different years: Five colonization and three infection strains recovered in 1980 and 1981; four colonization strains in 1990; seven infection and one colonization strains in 2000, and 10 infection strains in 2010. The intestinal colonization strains, isolated in 1990 and 2000, were obtained from a primary streak in Eosin Methylene Blue Agar medium (EMB agar - Difco Laboratories, Detroit, MI) containing 8 µg/ml of gentamicin. The intestinal colonization strains and the infection strains recovered in 1980 and 1981 were isolated and identified by the hospital bacte riology laboratory.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing (AST) was per formed using the agar diffusion method as set by the Clinical Laboratories Standards Institute (CLSI) (12). Escherichia coli strain ATCC 25922 was used as control, and the following antimicrobial agents were tested (suppliers´ individual concentrations appear in brackets): sulfamethaxazole - trimethropim (St- 23.75-1.25 µg), cephalotin (Cp- 30 µg), ceftazidime (Cz- 30 µg), cefoxitin (Fx- 30 µg), cefotaxime (Ct- 30 µg), cephazoline (Cf- 30 µg), chloranphenicol (Ch- 30 µg), ciprofloxacin (Ci- 5 µg), norfloxacin (Nr- 10 µg), tetracycline (Tt- 30 µg) and ampicillin (Ap- 10 µg). Aminoglycosides tested were: gentamicin (Gn- 10 µg), amikacin (Ak- 30 µg), kanamycin (Kn- 30 µg), tobramiycin (Tb- 10 µg), neomycin (Ne- 30 µg) and netilmycin (Nt- 30 µg).
Strains resistant to second and third generation cephalosporins in the AST were subjected to confirmatory tests for extended-spectrum beta-lactamases (ESBL) production using the double-disc synergy and the approximation tests as established by the CLSI (12).
Bacterial strains were grouped based on the results obtained in the susceptibility tests.
Plasmid DNA extraction and agarose gel electrophoresis in wild bacterial strains
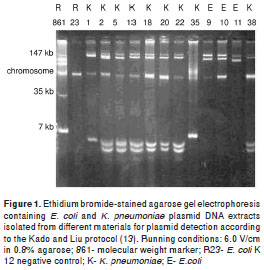
Twenty two of 30 strains were randomly selected and subjected to DNA extraction and electropho resis (in 0.8% agarose gel) for plasmid detection following the Kado and Liu protocol (13). We used the E. coli R861 strain plasmid DNA as molecular weight marker. Electrophoresis gels were dyed with ethidium bromide solution (0.5 µg/ml), analyzed in ultraviolet transluminator and photographed with a Kodak EDAS 120 system.
B2d DNA fragment cloning
The 1.9 Kb B2d DNA fragment (GenBank accession number GQ422439) was obtained from Kp 401F10 plasmid digestion with restriction enzymes. The plamid was obtained from the K. pneumoniae 20 Kp strain isolated from the faeces of a surgical patient in the 80´s. B2d fragment sequencing was also performed to use it as a positive control for PCR and a probe for DNA hybridization.
Automated DNA sequencing
B2d DNA fragment gene sequencing was done according to Otto, et al . (14). Sequences were ana lyzed and compared using the BioEdit Sequence Alignment Editor software (15).
PCR conditions
The thermal cycling conditions were performed in a Cetus model 480 thermal cycler (Perkin-Elmer, Norwalk, CT). The primers used were: aacC2 gene yielding a 237 bp product (5´-ACT GTG ATG GGA TAC GCG TC-3´ and 5´-CTC CGT CAG CGT TTC AGC TA-3´); aadB gene yelding a 320 bp product (5´-GAG CGA AAT CTG CCG CTC TGG-3´ and 5´- CTG TTA CAA CGG ACT GGC CGC-3´), and aacC3 gene yielding a 815 bp product (5´-AAA CTG GTG GCA ATA GAA GGA T-3´ and 5´- CTA TCC GTA TGA CGC TGA GTC3´), according to van de Klundert and Vliengenthart´s protocol (16). PCR assays were performed in a 50 m l total volume adding the following components to the reaction tubes: 1 µL of target DNA (obtained from dilutions of colonies in 50 m L of 10 mM Tris, 1 mM EDTA -pH 8.0), 1.5 mM MgCl 2 , 0.2 mM of dNTP mixture (dATP, dTTP, dCTP and dGTP), 20 pmol of each primer, 1x PCR buffer, and 1.25U of Taq DNA Polymerase. The amplified products were subjected to electrophoresis in a 2% agarose gel.
DNA hybridization assays
For the DNA hybridization assays we used Thomas protocol (17): After electrophoresis in agarose gel, DNA extracted from K. pneumoniae and E. coli wild strains was transferred to a nylon membrane by capillarity system. The 1.9 Kb B2d DNA fragment was labeled with a [dATP] P 32 through random primed labeling (Gibco®, Life Technologies, US) and used as probe according to Feinberg and Vogelstein´s method (18).
Results
Identifying the antimicrobial susceptibility profiles
Antimicrobial resistance profiles were determined based on the results of the AST performed with aminoglycosides and other drugs. K. pneumoniae and E. coli strains AST for aminoglycosides indicated a high frequency of resistance to Gn, Kn and Tb (21 strains: 70%). Regarding the other antimicrobial agents, AST indicated a high frequency of resistance to Cp, Ch, Tt and Ap (17 strains: 57%) (table 1).
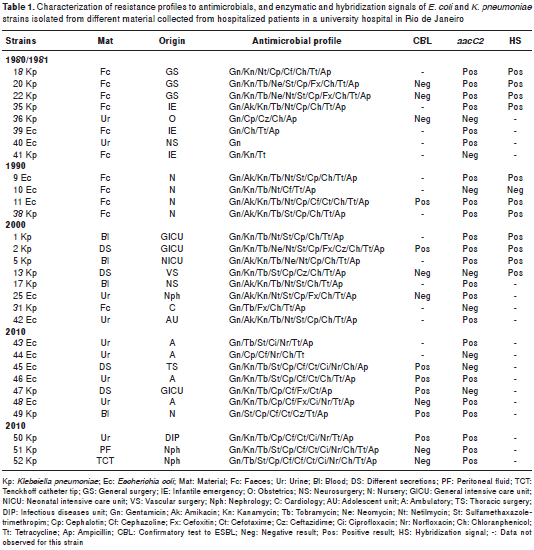
Fifteen strains were subjected to ESBL-production confirmatory tests and seven (46.7%) presented the ESBL phenotype. ESBL positive strains dis tribution was as follows: 1 (25%) from strains isolated in 1990; 1 (12.5%) from strains isolated in 2000, and 5 (50%) from strains isolated in 2010 (table 1). None of the strains isolated in 1980 or 1981 were ESBL positive. However, one strain (12.5%) isolated in this latter period, as well as one strain (12.5%) isolated in 2000, showed resistance to the third generation cephalosporin Cz (table 1).
Plasmid profile of strains
Out of the 22 strains randomly selected , 14 corresponded to K. pneumoniae and eigth to E. coli. They were subjected to plasmid DNA extractions with the following results: Nine strains (41%) had plasmids with molecular weights of 147 kb or more, and plasmids of around 7 kb (figure 1). Strains 9 Ec, 10 Ec and 11 Ec isolated in 1990 had similar eletrophoretic patterns, as well as strains 18 Kp , 20 Kp and 22 Kp isolated in 1980 and 1981, while only strain 38 Kp from 1990 and strains 2 Kp, 5 Kp and 13 Kp from 2000 showed such similarity.
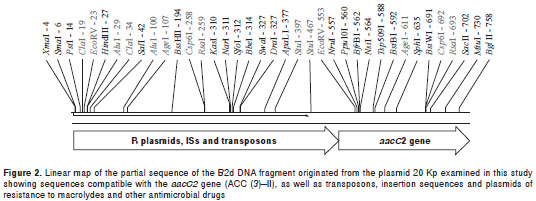
Analysis of DNA sequences
DNA sequencing was performed for the B2d DNA fragment originated from plasmid 20 Kp. The 1.9 kb repeat sequence obtained indicated the presence of DNA sequences homologous to the aacC2 gene or to the aminoglycoside-(3)-N-acetyltransferase (AAC (3) II). The BLAST TM analysis of the B2d fragment allowed us to identify a subset of repeats that showed over 90% of overall identity and E values <0.05 when compared to other GenBank records, mostly related to acetyltransferases, insertion sequences and transposons found in Gram-negative bacteria (figure 2). The most signi ficant sequences aligned to the B2d fragment were: Salmonella enterica subsp. enterica serovar Choleraesuis TnpA_rve (GI: 161867979); Esche richia coli IAI39 transposase, IS26 (GI: 218700153 ); Acinetobacter baumannii AB0057 transposase IS26 (GI: 213155651); Escherichia coli AAC(3)-II (GI: 41056930), and Citrobacter freundii aminoglycoside acetyltransferase (GI: 27383509).
Detection of sequences coding for aminoglycoside modifying enzymes
Amplification products indicated the presence of the aacC2 gene (GenBank access number X51534) in 13 (72,2%) K. pneumoniae strains and nine (75%) E. coli strains (table 1). We did not detect amplification for the other genes under study.
Hybridization profiles
Plasmid DNA was extracted from 12 strains which were subsequently subjected to DNA hybridization assays using the B2d fragment as probe (data not shown). Positive signals were obtained for 11 (91.7%) of the 12 strains tested (table 1). Regarding the extracts of Kp 18 and Kp 38 strains isolated in 1981 and 1990, respectively, hybridization signals were detected in the chromosome. Strain Kp 5, isolated in 2000, and strain Ec 11, isolated in 1990, showed more than one band, indicating that the gene sequences identified in the B2d fragment were present in several regions of the bacterial genome. In the other strains with more than one plasmid, the hybridization signal was detected in just one of the plasmid bands.
Discussion
To characterize the resistance profile of the isolated strains, we selected different classes of antimicrobial agents: those which have been used for a long time, as well as some classes introduced more recently in therapeutics, such as beta-lactam antibiotics and aminoglycosides , important antimicrobial drugs used in hospital environments to which microorganisms have increasingly become resistant (19,20).
ESBL- producing Gram-negative bacteria have been detected since the 1980s as important causes of nosocomial infections (21). After the introduction of expanded-spectrum cephalosporins, K. pneumoniae and E. coli resistant strains emerged in certain areas of the world (22). In fact, this study detected multidrug-resistant, aminoglycoside resistant and ESBL-producing strains in the different decades. Among those isolated in the 80´s, despite the absence of ESBL-producing strains, we detected resistance to third generation cephalosporins.
Aminoglycosides susceptibility profiles in strains isolated between September 1980 and January 1981 were similar to those in strains isolated in 1990, 2000 and 2010. TSA results agreed with those described in the literature for the acetylase identified as AAC(3)-II (Gen/Tob/Net) in most of the strains (8). Small changes in DNA sequences in genes throughout the replication processes, and/ or acquired genes, may have resulted in these phenotypical differences.
Several environmental and nosocomial bacteria can contain transferable plasmids that carry genes expressing several traits, one of which is multidrug-resistance to antimicrobial drugs (23-25). In fact, we found plasmids with molecular weights varying between 147 kb or more and 7 kb or less in multidrug-resistant strains isolated from especimens of patients hospitalized in different units and in different decades. These genetic elements can be transferred from a microorganism to another in vivo , vertically or horizontally, contributing to worsen hospital infections (3).
The similarity of plasmid profiles in strains isolated in different decades indicates the persistence of bacterial clones and/or of specific plasmids. Well established populations of bacterial clones can extend their resistance phenotypes by acquiring genetic elements (plasmids, integrons and transposons), facilitating co-selection processes under different levels of pressure and favoring the permanence of these microorganisms and their genetic constituents in the environment (26). The possibility of co-transmission may not only contribute to increase resistance markers, but also confer an evolutionary benefit to these strains, leading to selection in an environment with persistent antibiotic pressure by beta-lactams, aminoglycosides and quinolones (3,24,27).
The B2d clone sequencing showed that, besides sequences compatible with the aacC2 gene (ACC (3)–II), there are other sequences compatible with transposons, insertion sequences and plasmid- mediat ed resistance to macrolides and other anti-microbial drugs. With respect to the presence of more than one hybridization signal observed in two strains (Kp 5 and Ec 11), we considered the possibility of different insertion events occurring throughout several generations of bacteria, which may have involved different plasmids in the same bacterial cell. We observed how these DNA sequences can determine DNA insertions in one or more genomic sites, originating resistant strains, irrespective of the presence of original mobile genetic elements. When we compared sequence frames with those already described in the GenBank, we were able to detect identity regions in microorganisms from different species, indicating the possibility of horizontal resistance gene transfer involving plasmids, transposons and insertion sequences. When analyzing 160 strains of gentamicin-resistant E. coli isolated from human and animal samples, Ho, et al . (28), identified the aacC2 gene in 81.3% of them, thus confirming this possibility.
Based on the results obtained with the hybridiza tion and PCR assays, it can be said that genetic elements housing multidrug-resistance encoding sequences presumably remain in circulation among microorganisms in hospital environments for long periods of time, and that these microorganisms may colonize and cause infections in hospitalized patients. Isolated strains were tested not only for the same period as the Kp 20 strain from which the fragment used as probe was extracted (1980/81), but in those isolated 10, 20 and even 30 years later (1990, 2000 and 2010, respectively). Selective pressure can be acting in favor of the persist ence of several genes grouped in one operon that, otherwise, could be eliminated. Besides, it is important to note that gene cassettes can contain many of the resistance genes expressed in Gram-negative bacteria (6).
The Kp 20 strain was originally isolated from faeces of a patient admitted in a general surgery ward. It is known that intestinal microbiota components are affected during antimicrobial therapeutics decreasing their number, and favoring proliferation of opportunistic microorganisms which can disseminate and cause infections (29). The human intestine provides an important reservoir for multidrug-resistant Gram-negative bacteria, including Enterobacteriaceae species involved in infectious processes both in community and nosocomial environments, and the use of anti-microbial agents is one of the important factors for the selection of multidrug-resistant microorganisms (20,30).
We want to emphasize the importance of reducing hospital stays, as well as of a more discerning and conscious use of antimicrobial agents in inpatient and outpatient hospital environments, and of educational programs aimed at updating healthcare professionals, especially on the importance of adequate hand washing before and after patient care, to name only some of the measures to control the persistent dissemination of microorganisms with resistance genes and plasmids.
Acknowledgments
We are grateful to Fiocruz/PDTIS/Brazil for DNA sequencing and to Dr. Julio Cesar Delgado Correal (Hospital Universitário Pedro Ernesto HUPE/UERJ) for the Spanish version of the abstract.
Financial support
This research received support from grants of the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil.
Conflict of interest
The authors declare no conficts of interest.
Corresponding author: Verônica Dias Gonçalves, Departamento de Microbiologia, Inmunologia e Parasitologia, Faculdade de Ciências Médicas, Universidade do Estado do Rio de Janeiro, Av. Professor Manuel de Abreu, 444/ 3º andar, CEP: 20550-170, Rio de Janeiro, RJ, Brasil Telephone: (5521) 2868 8280; fax: (5521) 2868 8376 kaiura@bol.com.br
References
1. Speller DC. Hospital Associated Infection. In: Parker MT, Coller LH, editors. Topley and Wilson´s principles of bacteriology, virology and immunology. London: Edward Arnold; 1990. p. 141-71. [ Links ]
2. Ruef C. Nosocomial infections - Multiple fields of activity. Infection. 2000;28:339-40 . http://dx.doi.org/10.1007/s150100070001 [ Links ]
3. Stokes HW, Gillings MR. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram -negative pathogens. FEMS Microbiol Rev. 2011;35:790-819. http://dx.doi.org/10.1111/j.1574-6976.2011.00273.x [ Links ]
4. Carattoli A. Resistance plasmid families in Enterobac teriaceae. Antimicrob Agents Chemother. 2009;53:2227-38. http://dx.doi.org/10.1128/AAC.01707-08 [ Links ]
5. Martínez LJ, Baquero F. Interations among strategies associated with bacterial infection: Pathogenicity, epidemicity, and antibiotic resistance. Clin Microbiol Rev. 2002;15:647-79. http://dx.doi.org/10.1128/CMR.15.4.647-679.2002 [ Links ]
6. Giedraitiene A, Vitkauskiene A, Naginiene R, Pavilonis A. Antibiotic resistance mechanisms of clinically important bacteria review. Medicina (Kaunas). 2011;47:137-46. [ Links ]
7. Queiroz ML, Antunes P, Mourão J, Merquior VL, Machado E, Peixe LV. Characterization of extended- spectrum beta-lactamases, antimicrobial resistancegenes, and plasmid content in Escherichia coli isolates from differ ent sources in Rio de Janeiro, Brazil. Diagn Microbiol Infect Dis. 2012;74:91-4. http://dx.doi.org/10.1016/j.diagmicrobio.2012.05.019 [ Links ]
8. Lindemann PC, Risberg K, Wilker HG, Mylvaganam H. Aminoglycoside resistance in clinical Escherichia coli and Klebsiella pneumoniae isolates from Western Norway. APMIS. 2011;120:495-502. http://dx.doi.org/10.1111/j.1600-0463.2011.02856.x [ Links ]
9. Ramírez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13:151-71. http://dx.doi.org/10.1016/j.drup.2010.08.003 [ Links ]
10. Barros JC, Pinheiro SR, Bozza M, Gueiros-Filho FJ, Bello AB, Lopes UG, et al . Evidences of gentamicin resistance amplification in Klebsiella pneumoniae isolated from faeces of hospitalized newborns. Mem Inst Oswaldo Cruz. 1999;94:795-802. http://dx.doi.org/10.1590/S0074-02761999000600016 [ Links ]
11. Gonçalves VD. Detecção e caracterização de enzimas modificadoras de aminoglicosídeos em cepas de enterobactérias multirresistentes de origem hospitalar [dissertação]. Rio de Janeiro: Universidade do Estado do Rio de Janeiro; 2001. [ Links ]
12. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; Twentieth informational supplement. Wayne, PA: CLSI; 2012. [ Links ]
13. Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365-73. [ Links ]
14. Otto TD, Vasconcellos EA, Gomes LH, Moreira AS, Degrave WM, Mendonça-Lima L, et al . ChromaPipe: A pipeline for analysis, quality control and management for a DNA sequencing facility. Genet Mol Res. 2008;7:861-71. [ Links ]
15. Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95-8. [ Links ]
16. van de Klundert JAM, Vliengenthart JS. PCR detection of genes coding aminoglicoside-modifying enzymes. In: Persing DH, Smth TF, Tenover FC, White TJ, editors. Diagnostic molecular microbiology: Principles and appli cations. Rochester: Mayo Foudation; 1993. p. 547-52. [ Links ]
17. Thomas PS. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci USA. 1980;77:5201-5. [ Links ]
18. Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6-13. http://dx.doi.org/10.1016/0003-2697(83)90418-9 [ Links ]
19. Yan JJ, Wu JJ, Ko WC, Tsai SH, Chuang CL, Wu HM, et al . Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J Antimicrob Chemother. 2004;54:1007-12. http://dx.doi.org/10.1093/jac/dkh455 [ Links ]
20. Donskey CJ. Antibiotic regimens and intestinal colonization with antibiotic-resistant gram-negative bacilli. Clin Infect Dis . 2006; 43(Suppl.2):S62-9. http://dx.doi.org/10.1086/504481 [ Links ]
21. Gniadkowski M. Evolution and epidemiology of extended- spectrum beta-lactamases (ESBLs) and ESBL-producing microorganisms. Clin Microbiol Infect. 2001;7:597-608. http://dx.doi.org/10.1046/j.1198-743x.2001.00330.x [ Links ]
22. Thomson KS, Prevan AM, Sanders CC. Novel plasmid- mediated beta-lactamases in enterobacteriaceae: Emerging problems for new B-lactam antibiotics. Curr Clin Topics Infect Dis. 1996;16:151-63. [ Links ]
23. de Meirelles-Pereira F, Meirelles AS, Gomes-da-Silva MC, Gonçalves VD, Brum PR, Castro EA, et al . Ecological aspects of the antimicrobial resistance in bacteria of impor tance to human infections. Braz J Microbiol. 2002;33:287- 93. http://dx.doi.org/10.1590/S1517-83822002000400002 [ Links ]
24. Ma L, Lin CJ, Chen JH, Fung CP, Chang FY, Lai YK, et al . Widespread dissemination of aminoglycoside resistance genes armA and rmtB in Klebsiella pneumoniae isolates in Taiwan producing CTX-M-type extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 2009;53:104-11. http://dx.doi.org/10.1128/AAC.00852-08 [ Links ]
25. Sun H, Li S, Xie Z, Yang F, Sun Y, Zhuhia Y, et al . A novel multidrug resistance plasmid isolated from a Escherichia coli strain resistant to aminoglycosides. J Antimicrob Chemother. 2012;67:1635-8. http://dx.doi.org/10.1093/jac/dks107 [ Links ]
26. Canton R, Loza E, Pascual A, Tubal F, Morosini MI, Almaraz F, et al . Comparative in vitro activity of garenoxacin (BMS 284756). Sentry program, Spain (1990- 2000). Enferm Infecc Microbiol Clin. 2003;21:404-6. [ Links ]
27. Wener KM, Schechner V, Gold HS, Wright SB, Carmeli Y. Treatment with fuoroquinolones or with ß -lactam- ß - lactamase inhibitor combinations is a risk factor for isolation of extended-spectrum- ß -lactamase-producing Klebsiella species in hospitalized patients. Antimicrob Agents Chemother. 2010;54:2010-6. http://dx.doi.org/10.1128/AAC.01131-09 [ Links ]
28. Ho P, Wong RC, Lo RW, Chow H, Wong SS, Que T. Genetic identity of aminoglycoside-resistance genes in Escherichia coli isolates from human and animal sources. J Med Microbiol. 2010;59:702-7. http://dx.doi.org/10.1099/jmm.0.015032-0 [ Links ]
29. Guzmán-Blanco M, Casellas JM, Sader HS. Bacterial resistance to antimicrobial agents in Latin America: The giant is awakening. Infect Dis Clin North Am. 2000;14:67-79. [ Links ]
30. Pereira JA, Suassuna I. Relações epidemiológicas entre Klebsiella pneumoniae isoladas de fezes e de infecções urinárias hospitalares. Rev Latinoam Microbiol. 1986;28:201-9. [ Links ]













